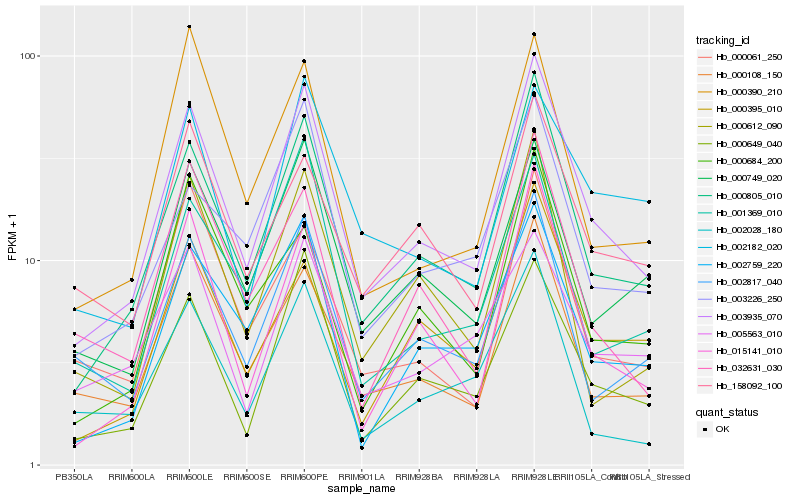
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003935_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000805_010 |
0.0578995774 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 3 |
Hb_000649_040 |
0.1045571068 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 4 |
Hb_000108_150 |
0.1112781849 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 5 |
Hb_015141_010 |
0.1144312189 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 6 |
Hb_000749_020 |
0.1182261433 |
- |
- |
Nucleotide-diphospho-sugar transferases superfamily protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_002759_220 |
0.1203440757 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 8 |
Hb_002817_040 |
0.1215396066 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 9 |
Hb_000612_090 |
0.1316819841 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 10 |
Hb_000395_010 |
0.1330906219 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 11 |
Hb_003226_250 |
0.1353093544 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 12 |
Hb_032631_030 |
0.1356966752 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 13 |
Hb_158092_100 |
0.1361857105 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 14 |
Hb_002028_180 |
0.1388037719 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_000390_210 |
0.1416366957 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 16 |
Hb_000061_250 |
0.1418522682 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 17 |
Hb_001369_010 |
0.1419075441 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 18 |
Hb_002182_020 |
0.1435807376 |
- |
- |
Thylakoid lumenal 17.4 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000684_200 |
0.1438312391 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 20 |
Hb_005563_010 |
0.144537117 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |