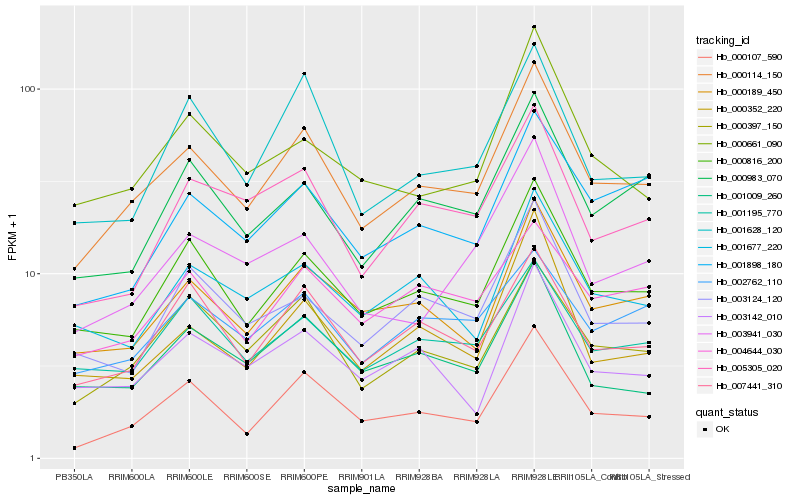
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000114_150 |
0.0 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_000816_200 |
0.0790049291 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 3 |
Hb_000107_590 |
0.0845703687 |
- |
- |
PREDICTED: tricalbin-3 [Jatropha curcas] |
| 4 |
Hb_000397_150 |
0.0852243008 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 5 |
Hb_000189_450 |
0.0896223815 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_007441_310 |
0.0962857715 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000983_070 |
0.1016393061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005305_020 |
0.1033597797 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001195_770 |
0.1056307517 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 10 |
Hb_001628_120 |
0.1066369068 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 11 |
Hb_001009_260 |
0.1068689618 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003124_120 |
0.1103042059 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 13 |
Hb_001898_180 |
0.1111679913 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001677_220 |
0.1119954646 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 15 |
Hb_004644_030 |
0.1128189185 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 16 |
Hb_000661_090 |
0.1133496716 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002762_110 |
0.1140967046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000352_220 |
0.1152316151 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_003142_010 |
0.11621743 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 20 |
Hb_003941_030 |
0.1172359474 |
- |
- |
Protease ecfE, putative [Ricinus communis] |