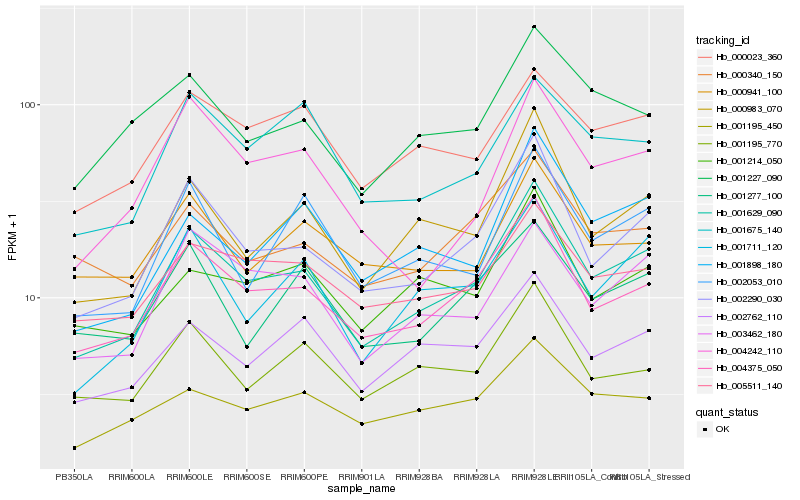
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001629_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002290_030 |
0.0606402279 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004375_050 |
0.0642071308 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000983_070 |
0.0856391279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001711_120 |
0.0927980046 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003462_180 |
0.0972680092 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 7 |
Hb_002762_110 |
0.1028654245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001898_180 |
0.1030220576 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005511_140 |
0.1059921372 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 10 |
Hb_000023_360 |
0.1077643478 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002053_010 |
0.1080233145 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_001214_050 |
0.1089157916 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001195_450 |
0.1089975215 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 14 |
Hb_001227_090 |
0.1090072062 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 15 |
Hb_001195_770 |
0.1095870959 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 16 |
Hb_000340_150 |
0.1115306017 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 17 |
Hb_001675_140 |
0.1137725058 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 18 |
Hb_001277_100 |
0.1142951126 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 19 |
Hb_004242_110 |
0.1166288646 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 20 |
Hb_000941_100 |
0.1190488646 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |