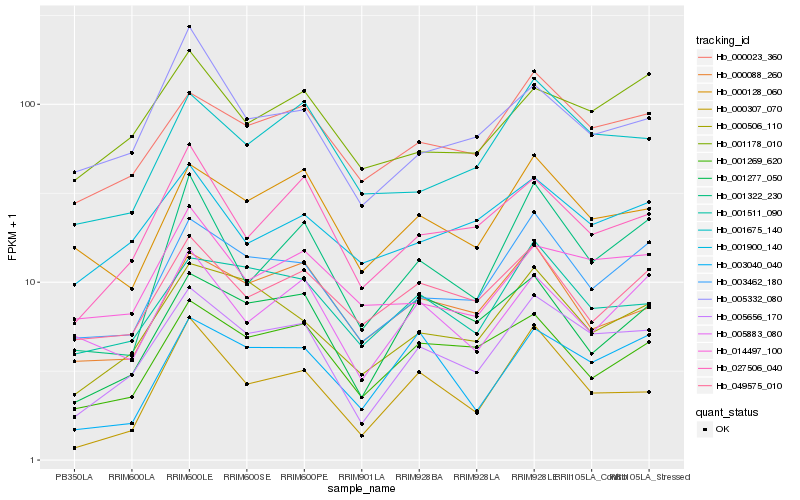
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005656_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637447 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003462_180 |
0.1008192578 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 3 |
Hb_000506_110 |
0.1071079328 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 4 |
Hb_000023_360 |
0.1083947327 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_027506_040 |
0.1135120145 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001511_090 |
0.1184094923 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 7 |
Hb_001277_050 |
0.1226252906 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001675_140 |
0.1233279518 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 9 |
Hb_001269_620 |
0.1248834606 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 10 |
Hb_000128_060 |
0.1268827534 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 11 |
Hb_000307_070 |
0.1305387129 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001322_230 |
0.1311548357 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 13 |
Hb_001178_010 |
0.1331850437 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 14 |
Hb_000088_260 |
0.134083029 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 15 |
Hb_003040_040 |
0.1349265035 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_014497_100 |
0.1374208073 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 17 |
Hb_005332_080 |
0.1383986317 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001900_140 |
0.1389387137 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005883_080 |
0.1389404879 |
- |
- |
type 2A protein phosphatase-2 [Populus trichocarpa] |
| 20 |
Hb_049575_010 |
0.1394013159 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |