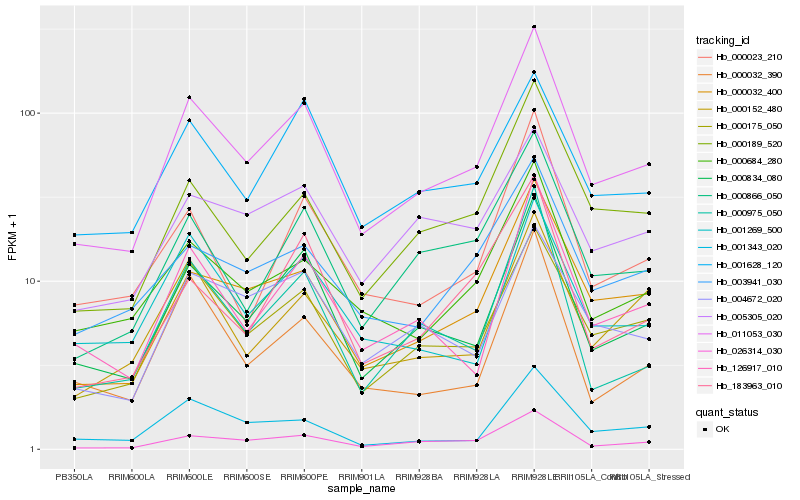
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011053_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000834_080 |
0.0706289444 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 3 |
Hb_000175_050 |
0.0887926246 |
- |
- |
- |
| 4 |
Hb_000032_400 |
0.0897750365 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 5 |
Hb_026314_030 |
0.0917511746 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 6 |
Hb_000866_050 |
0.1056872645 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_126917_010 |
0.1077904413 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 8 |
Hb_000684_280 |
0.1082974583 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000189_520 |
0.114229999 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 10 |
Hb_003941_030 |
0.1192453921 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 11 |
Hb_001628_120 |
0.1223917519 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 12 |
Hb_000975_050 |
0.1229182625 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_183963_010 |
0.1239449478 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 14 |
Hb_000152_480 |
0.1241668269 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004672_020 |
0.1241830404 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 16 |
Hb_000023_210 |
0.1248908826 |
- |
- |
PREDICTED: protein TIC 55, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001343_020 |
0.1259647306 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000032_390 |
0.1279417278 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 19 |
Hb_001269_500 |
0.1288462621 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 20 |
Hb_005305_020 |
0.129166411 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |