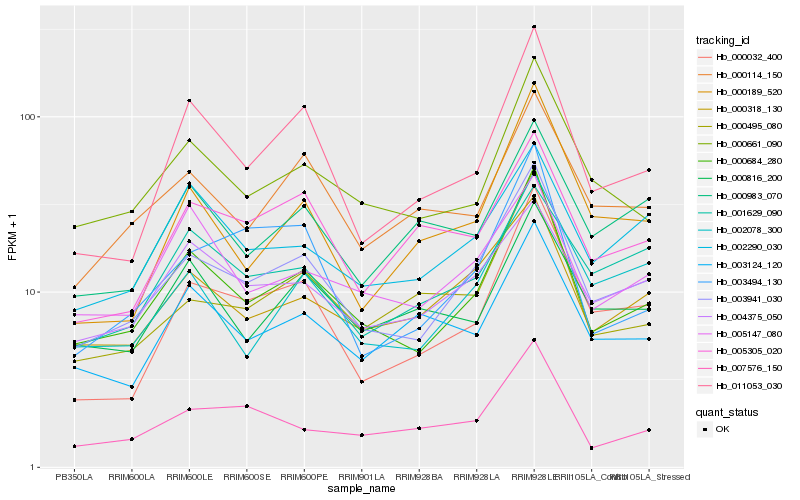
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003941_030 |
0.0 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 2 |
Hb_000684_280 |
0.0681347784 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000318_130 |
0.0998487919 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000032_400 |
0.1054745363 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 5 |
Hb_000661_090 |
0.1134922577 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002078_300 |
0.1148558681 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004375_050 |
0.115693417 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000114_150 |
0.1172359474 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_011053_030 |
0.1192453921 |
- |
- |
- |
| 10 |
Hb_002290_030 |
0.1247712771 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000495_080 |
0.1276720057 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 12 |
Hb_001629_090 |
0.1306649367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005305_020 |
0.1312448096 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_007576_150 |
0.1335938312 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g57430, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003494_130 |
0.1336457895 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 16 |
Hb_000189_520 |
0.1339973468 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 17 |
Hb_000983_070 |
0.1345678756 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000816_200 |
0.1351946663 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 19 |
Hb_003124_120 |
0.1365729418 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 20 |
Hb_005147_080 |
0.1379233004 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |