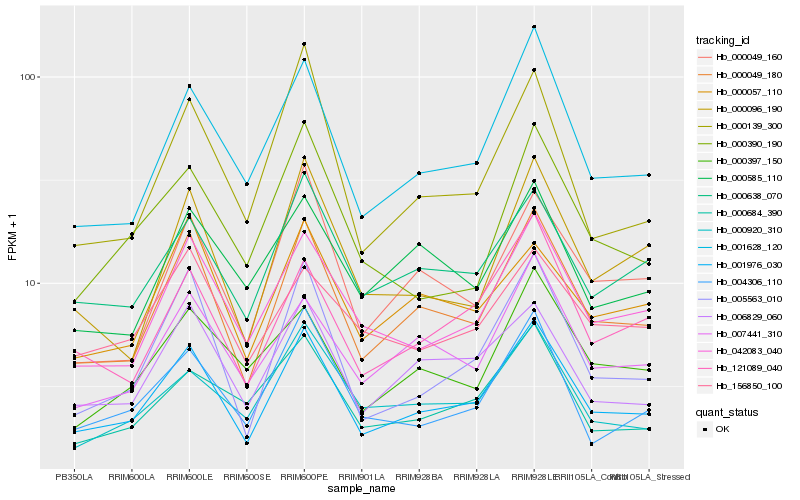
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001976_030 |
0.0 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000049_180 |
0.0477540273 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 3 |
Hb_001628_120 |
0.0792784707 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 4 |
Hb_042083_040 |
0.0842885639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006829_060 |
0.0877609761 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005563_010 |
0.0908330588 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 7 |
Hb_000638_070 |
0.0921320035 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 8 |
Hb_121089_040 |
0.0948329214 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 9 |
Hb_004306_110 |
0.0985459197 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 10 |
Hb_007441_310 |
0.099031011 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000920_310 |
0.1051102185 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 12 |
Hb_000096_190 |
0.1067478451 |
- |
- |
unknown [Lotus japonicus] |
| 13 |
Hb_000049_160 |
0.1081802319 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 14 |
Hb_000684_390 |
0.1093094141 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 15 |
Hb_000139_300 |
0.1100604118 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 16 |
Hb_000057_110 |
0.1106552894 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 17 |
Hb_000390_190 |
0.1121217311 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000397_150 |
0.1135875683 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 19 |
Hb_156850_100 |
0.1151024105 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 20 |
Hb_000585_110 |
0.1167755139 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |