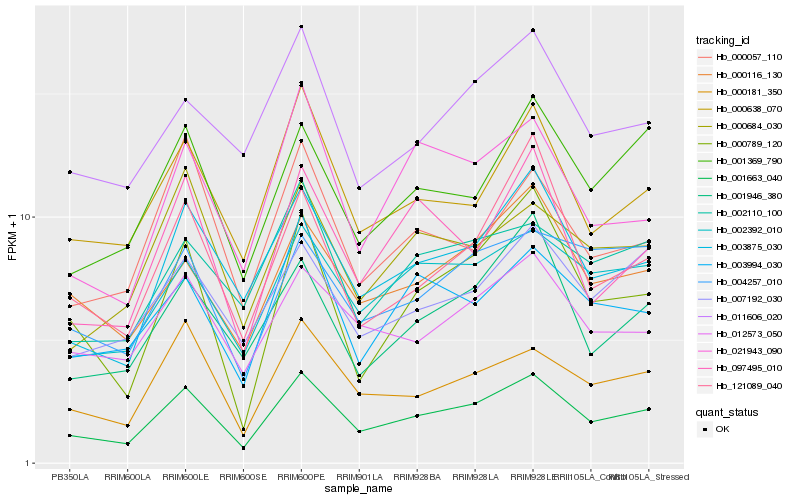
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001663_040 |
0.0 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 2 |
Hb_003875_030 |
0.0840133502 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 3 |
Hb_000181_350 |
0.0875160032 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 4 |
Hb_000638_070 |
0.0924346151 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 5 |
Hb_002392_010 |
0.0954118031 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 6 |
Hb_012573_050 |
0.0977539555 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 7 |
Hb_000116_130 |
0.0979210508 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 8 |
Hb_001369_790 |
0.1027423007 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003994_030 |
0.1037050573 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 10 |
Hb_000789_120 |
0.1070097978 |
- |
- |
PREDICTED: derlin-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_021943_090 |
0.1073282251 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 12 |
Hb_097495_010 |
0.1074523205 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007192_030 |
0.1082817393 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 14 |
Hb_000684_030 |
0.1082847942 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 15 |
Hb_001946_380 |
0.1085869227 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 16 |
Hb_011606_020 |
0.1096465571 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 17 |
Hb_121089_040 |
0.1096547486 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 18 |
Hb_002110_100 |
0.1101548284 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000057_110 |
0.1114326755 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 20 |
Hb_004257_010 |
0.1135600018 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |