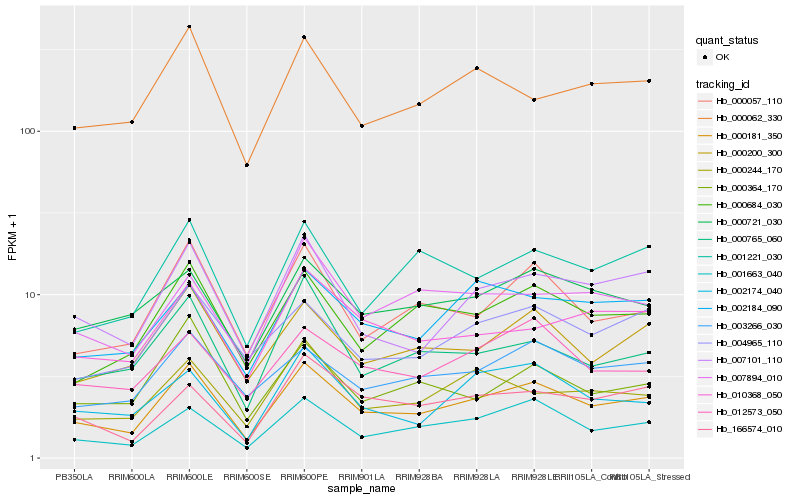
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_350 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 2 |
Hb_001663_040 |
0.0875160032 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 3 |
Hb_007101_110 |
0.0926029327 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 4 |
Hb_000057_110 |
0.0942248556 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 5 |
Hb_000200_300 |
0.0987277623 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 6 |
Hb_000684_030 |
0.100517145 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 7 |
Hb_000062_330 |
0.1104186108 |
- |
- |
unknown [Lotus japonicus] |
| 8 |
Hb_003266_030 |
0.1111555322 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 9 |
Hb_001221_030 |
0.111571102 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 10 |
Hb_004965_110 |
0.1118210554 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 11 |
Hb_010368_050 |
0.111895187 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 12 |
Hb_000244_170 |
0.1124827786 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 13 |
Hb_002184_090 |
0.1127854882 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_166574_010 |
0.1132876736 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 15 |
Hb_000765_060 |
0.1135139009 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 16 |
Hb_002174_040 |
0.1149801952 |
- |
- |
DNAJ heat shock family protein [Theobroma cacao] |
| 17 |
Hb_012573_050 |
0.1150197759 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 18 |
Hb_007894_010 |
0.1179659974 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 19 |
Hb_000721_030 |
0.1179768617 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 20 |
Hb_000364_170 |
0.1179921316 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |