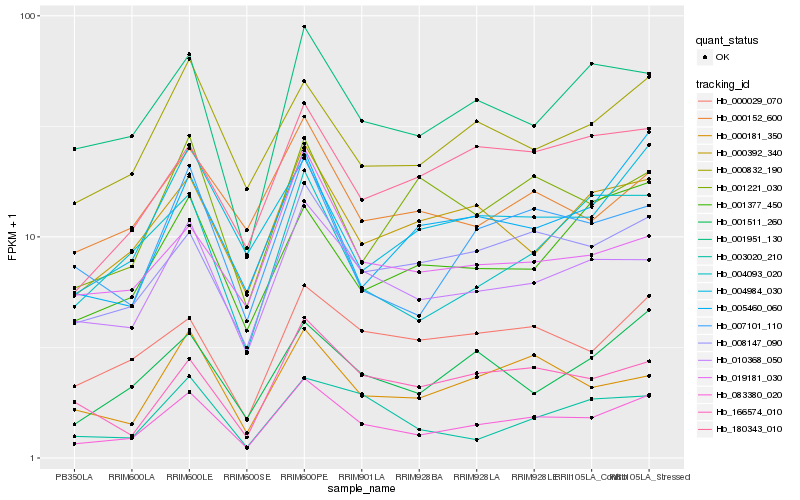
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083380_020 |
0.0 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 2 |
Hb_010368_050 |
0.1002133088 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 3 |
Hb_008147_090 |
0.1066635016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007101_110 |
0.1144326185 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 5 |
Hb_001951_130 |
0.1180634631 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 6 |
Hb_000181_350 |
0.1200565658 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 7 |
Hb_001511_260 |
0.1204922866 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 8 |
Hb_000832_190 |
0.1227915808 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 9 |
Hb_000029_070 |
0.1251736332 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 10 |
Hb_000392_340 |
0.1264663141 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |
| 11 |
Hb_004093_020 |
0.1267001664 |
- |
- |
Calcium-binding EF-hand family protein [Theobroma cacao] |
| 12 |
Hb_166574_010 |
0.1267437513 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 13 |
Hb_004984_030 |
0.1280495023 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_005460_060 |
0.1309626296 |
- |
- |
PREDICTED: proteasome assembly chaperone 2 [Jatropha curcas] |
| 15 |
Hb_180343_010 |
0.134648817 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 16 |
Hb_019181_030 |
0.1369459285 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 17 |
Hb_003020_210 |
0.1369546542 |
- |
- |
Structural maintenance of chromosomes 2-2 -like protein [Gossypium arboreum] |
| 18 |
Hb_001377_450 |
0.1372286927 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000152_600 |
0.1372553569 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 20 |
Hb_001221_030 |
0.1373531597 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |