| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
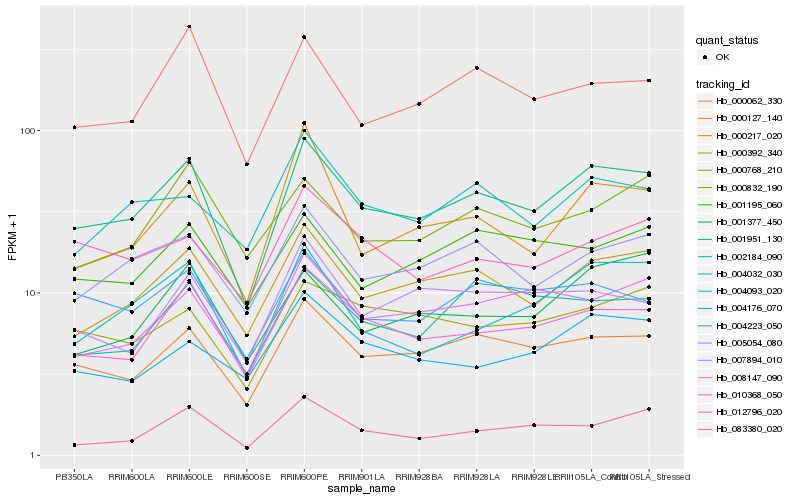
Hb_001951_130 |
0.0 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 2 |
Hb_000127_140 |
0.0794627379 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 3 |
Hb_010368_050 |
0.0817405014 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 4 |
Hb_000392_340 |
0.0825746692 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |
| 5 |
Hb_005054_080 |
0.0985717681 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 6 |
Hb_008147_090 |
0.1082506894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000062_330 |
0.1083259528 |
- |
- |
unknown [Lotus japonicus] |
| 8 |
Hb_004093_020 |
0.1091406344 |
- |
- |
Calcium-binding EF-hand family protein [Theobroma cacao] |
| 9 |
Hb_004223_050 |
0.1110905866 |
- |
- |
hexokinase [Manihot esculenta] |
| 10 |
Hb_002184_090 |
0.115265646 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000768_210 |
0.115641703 |
- |
- |
PREDICTED: uncharacterized protein LOC105632522 [Jatropha curcas] |
| 12 |
Hb_083380_020 |
0.1180634631 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 13 |
Hb_004032_030 |
0.1192083425 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 14 |
Hb_001195_060 |
0.1206743971 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 15 |
Hb_001377_450 |
0.1217603692 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000217_020 |
0.1224568848 |
- |
- |
AMME syndrome candidateprotein 1 protein, putative [Ricinus communis] |
| 17 |
Hb_004176_070 |
0.1229789598 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000832_190 |
0.1231801616 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 19 |
Hb_007894_010 |
0.1232938714 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 20 |
Hb_012796_020 |
0.1237002508 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 [Prunus mume] |