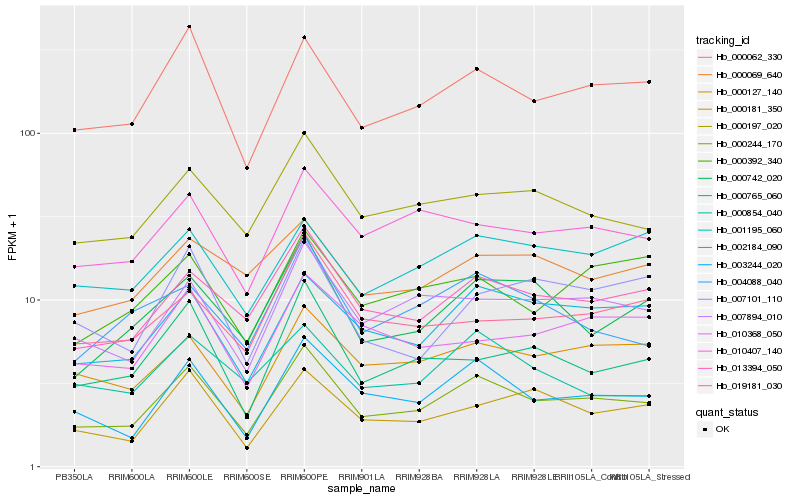
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000244_170 |
0.0 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 2 |
Hb_003244_020 |
0.0786935136 |
- |
- |
PREDICTED: serine carboxypeptidase-like 27 [Jatropha curcas] |
| 3 |
Hb_000062_330 |
0.0859389627 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_013394_050 |
0.0883685444 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |
| 5 |
Hb_007894_010 |
0.0971995184 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 6 |
Hb_002184_090 |
0.0998072584 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000197_020 |
0.1033420221 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 8 |
Hb_000127_140 |
0.1053737672 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 9 |
Hb_000742_020 |
0.1102923907 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 10 |
Hb_010407_140 |
0.1112405011 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 11 |
Hb_000765_060 |
0.111389477 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 12 |
Hb_000181_350 |
0.1124827786 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 13 |
Hb_010368_050 |
0.1203484633 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 14 |
Hb_004088_040 |
0.1213403966 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' iota isoform [Jatropha curcas] |
| 15 |
Hb_000392_340 |
0.1247452946 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |
| 16 |
Hb_019181_030 |
0.1250651012 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 17 |
Hb_007101_110 |
0.125941946 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 18 |
Hb_000069_640 |
0.1282246885 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 19 |
Hb_001195_060 |
0.1283284052 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 20 |
Hb_000854_040 |
0.129210635 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |