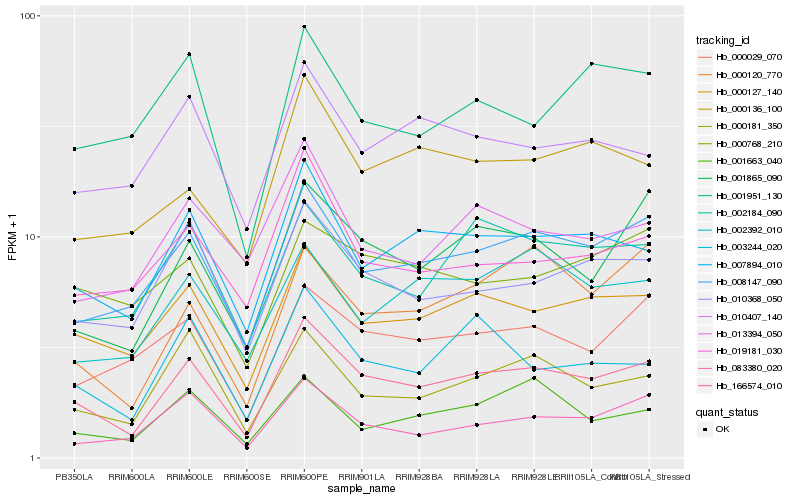
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_166574_010 |
0.0 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 2 |
Hb_000127_140 |
0.0939952609 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 3 |
Hb_008147_090 |
0.0960868517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007894_010 |
0.0972480677 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 5 |
Hb_001865_090 |
0.1050111088 |
- |
- |
hypothetical protein VITISV_005773 [Vitis vinifera] |
| 6 |
Hb_010368_050 |
0.1106955134 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 7 |
Hb_000181_350 |
0.1132876736 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 8 |
Hb_003244_020 |
0.1191570194 |
- |
- |
PREDICTED: serine carboxypeptidase-like 27 [Jatropha curcas] |
| 9 |
Hb_002184_090 |
0.1206610304 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000029_070 |
0.1231448738 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 11 |
Hb_083380_020 |
0.1267437513 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 12 |
Hb_019181_030 |
0.1305183128 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 13 |
Hb_000136_100 |
0.1306434262 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 14 |
Hb_000120_770 |
0.1319218621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_010407_140 |
0.1337210825 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 16 |
Hb_001951_130 |
0.1349613699 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 17 |
Hb_001663_040 |
0.1353769438 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 18 |
Hb_000768_210 |
0.1359864564 |
- |
- |
PREDICTED: uncharacterized protein LOC105632522 [Jatropha curcas] |
| 19 |
Hb_013394_050 |
0.1365231487 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |
| 20 |
Hb_002392_010 |
0.1367343504 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |