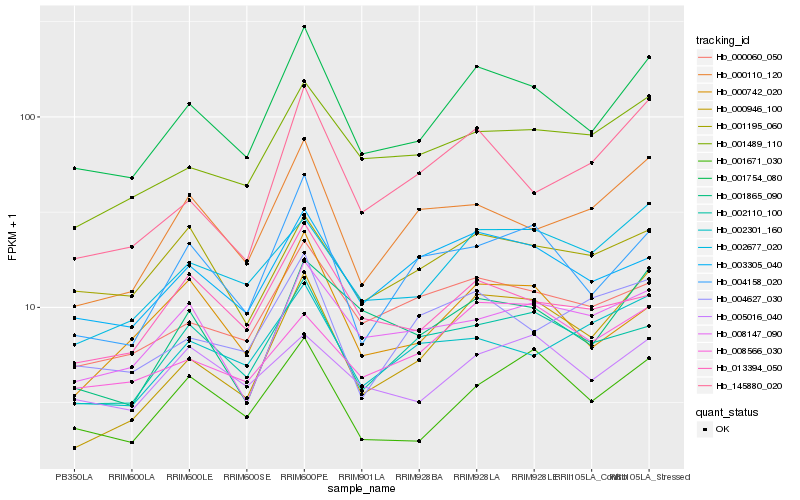
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_080 |
0.0 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 2 |
Hb_000060_050 |
0.1037380562 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 3 |
Hb_004158_020 |
0.1055542785 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 4 |
Hb_003305_040 |
0.1057315423 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 5 |
Hb_001489_110 |
0.1068521439 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Citrus sinensis] |
| 6 |
Hb_002110_100 |
0.1070577819 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002677_020 |
0.1073382622 |
- |
- |
PREDICTED: uncharacterized protein LOC105642265 [Jatropha curcas] |
| 8 |
Hb_013394_050 |
0.1113307902 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |
| 9 |
Hb_001865_090 |
0.1141694324 |
- |
- |
hypothetical protein VITISV_005773 [Vitis vinifera] |
| 10 |
Hb_001671_030 |
0.1143063766 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 11 |
Hb_000742_020 |
0.1168709002 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 12 |
Hb_000110_120 |
0.1176612625 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 13 |
Hb_008147_090 |
0.1188714471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_145880_020 |
0.1223945948 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 15 |
Hb_004627_030 |
0.1249080089 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 16 |
Hb_005016_040 |
0.1252186369 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 17 |
Hb_001195_060 |
0.1255298121 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 18 |
Hb_000946_100 |
0.126498768 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 19 |
Hb_002301_160 |
0.1267500072 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 20 |
Hb_008566_030 |
0.1270515729 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |