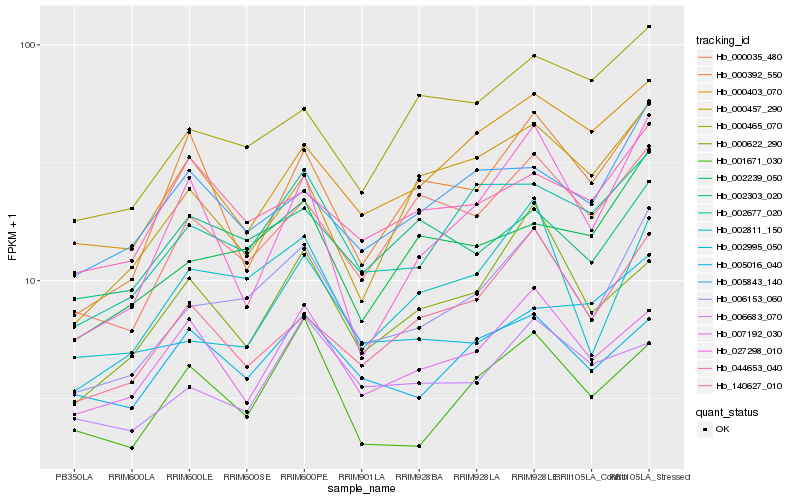
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006153_060 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_002811_150 |
0.0930833488 |
- |
- |
PREDICTED: uncharacterized protein LOC105647666 [Jatropha curcas] |
| 3 |
Hb_002677_020 |
0.1008701901 |
- |
- |
PREDICTED: uncharacterized protein LOC105642265 [Jatropha curcas] |
| 4 |
Hb_002239_050 |
0.1112284459 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000035_480 |
0.1168308375 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 6 |
Hb_140627_010 |
0.1224376612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000403_070 |
0.1275763309 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_007192_030 |
0.1306129836 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 9 |
Hb_005843_140 |
0.1319499725 |
- |
- |
Red chlorophyll catabolite reductase, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000465_070 |
0.1324978968 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase small subunit 3-like [Jatropha curcas] |
| 11 |
Hb_000622_290 |
0.1326214986 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 12 |
Hb_001671_030 |
0.1326345163 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 13 |
Hb_005016_040 |
0.1340247524 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 14 |
Hb_006683_070 |
0.1375353671 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 15 |
Hb_000457_290 |
0.1379464265 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 16 |
Hb_002303_020 |
0.1396194435 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Gossypium raimondii] |
| 17 |
Hb_044653_040 |
0.1410284906 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 18 |
Hb_002995_050 |
0.1413741548 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000392_550 |
0.1429707722 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_027298_010 |
0.1429778732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |