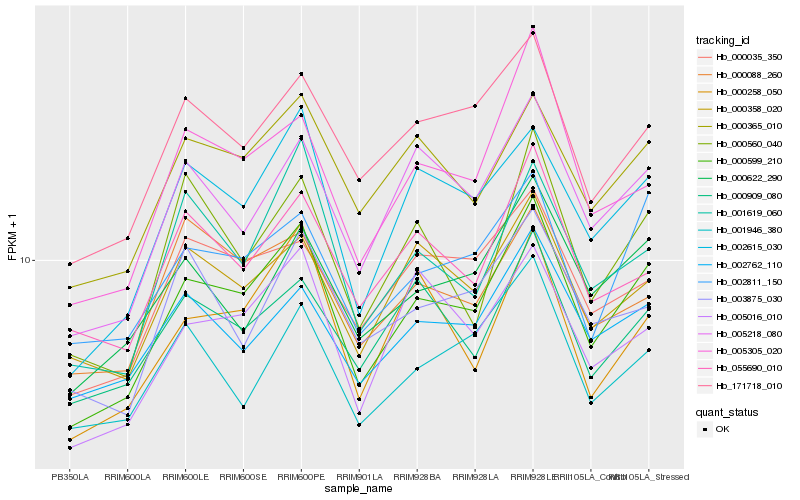
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_210 |
0.0 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 2 |
Hb_002615_030 |
0.0871883376 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 3 |
Hb_005218_080 |
0.0960809291 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_171718_010 |
0.0980814598 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 5 |
Hb_000365_010 |
0.0995054148 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 6 |
Hb_002811_150 |
0.1030360059 |
- |
- |
PREDICTED: uncharacterized protein LOC105647666 [Jatropha curcas] |
| 7 |
Hb_000622_290 |
0.1039794204 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 8 |
Hb_005305_020 |
0.1060696238 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000358_020 |
0.1085419491 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 10 |
Hb_002762_110 |
0.1089952016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005016_010 |
0.1094283038 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 12 |
Hb_001619_060 |
0.1105457836 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000035_350 |
0.1134311617 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000909_080 |
0.1139545361 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 15 |
Hb_055690_010 |
0.1139965694 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000258_050 |
0.1146422944 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000560_040 |
0.1157478348 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001946_380 |
0.1179312471 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 19 |
Hb_003875_030 |
0.1241020016 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 20 |
Hb_000088_260 |
0.1250082466 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |