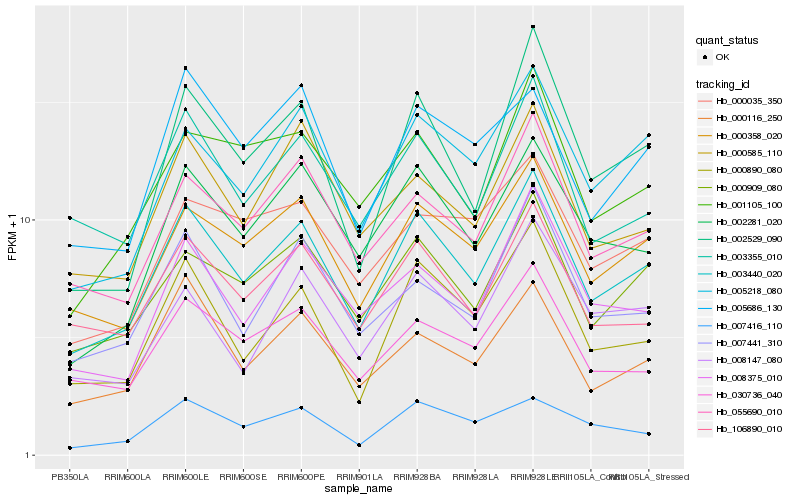
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003440_020 |
0.0 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_000358_020 |
0.0672546522 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 3 |
Hb_000909_080 |
0.0718762166 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 4 |
Hb_005218_080 |
0.0789463998 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_055690_010 |
0.0808234171 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000890_080 |
0.0835654071 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 7 |
Hb_106890_010 |
0.0837512024 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_002281_020 |
0.0840544291 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 9 |
Hb_030736_040 |
0.0863015147 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003355_010 |
0.0864379781 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 11 |
Hb_002529_090 |
0.089152784 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_000116_250 |
0.0899031869 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 13 |
Hb_008375_010 |
0.0937288209 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 14 |
Hb_000585_110 |
0.0953857853 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 15 |
Hb_001105_100 |
0.0965701517 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 16 |
Hb_007441_310 |
0.0967296598 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007416_110 |
0.1016139601 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008147_080 |
0.1018054226 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005686_130 |
0.1032656663 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 20 |
Hb_000035_350 |
0.1043715284 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |