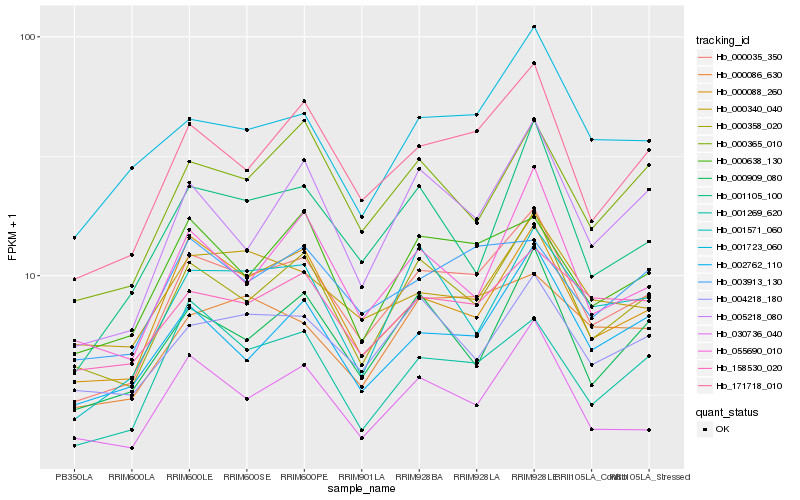
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_350 |
0.0 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_171718_010 |
0.0622988932 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 3 |
Hb_000358_020 |
0.0717616068 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 4 |
Hb_000088_260 |
0.0837205651 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 5 |
Hb_000638_130 |
0.0939008017 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 6 |
Hb_005218_080 |
0.0948378098 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_030736_040 |
0.0954559721 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002762_110 |
0.0957603515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001269_620 |
0.0968091483 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 10 |
Hb_001723_060 |
0.0987884812 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 11 |
Hb_000365_010 |
0.0991477353 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 12 |
Hb_003913_130 |
0.0993670894 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 13 |
Hb_000909_080 |
0.0996531961 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 14 |
Hb_055690_010 |
0.0996913113 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000086_630 |
0.0997532996 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_158530_020 |
0.1003112066 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 17 |
Hb_001105_100 |
0.1008338689 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 18 |
Hb_001571_060 |
0.1028440951 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 19 |
Hb_000340_040 |
0.1030169262 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 20 |
Hb_004218_180 |
0.1032935633 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |