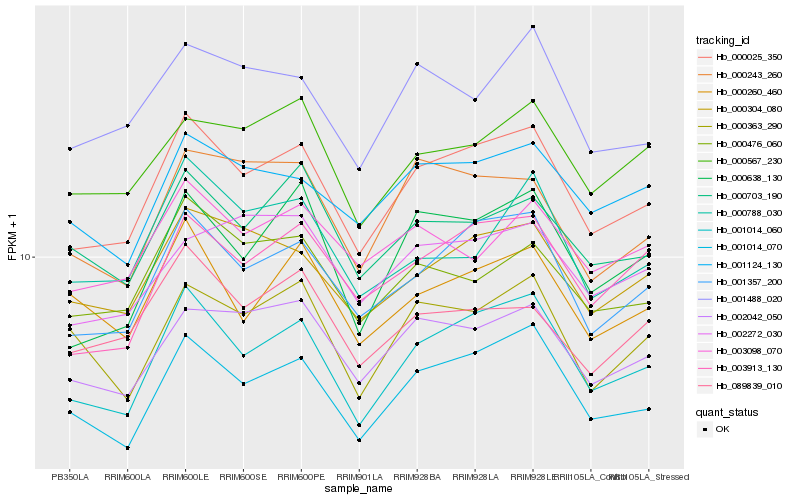
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_350 |
0.0 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001014_060 |
0.0392115559 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 3 |
Hb_001357_200 |
0.0419500729 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_001014_070 |
0.0515946376 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 5 |
Hb_003913_130 |
0.0681127516 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 6 |
Hb_000304_080 |
0.0681637989 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002042_050 |
0.0704081599 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000638_130 |
0.0712790044 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 9 |
Hb_001124_130 |
0.07619487 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 10 |
Hb_000476_060 |
0.0782414471 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_001488_020 |
0.0804492744 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 12 |
Hb_000567_230 |
0.0816286063 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 13 |
Hb_000703_190 |
0.0817838002 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000243_260 |
0.0823818516 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000260_460 |
0.0845079086 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000363_290 |
0.0856707696 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 17 |
Hb_003098_070 |
0.087216562 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 18 |
Hb_089839_010 |
0.0873360668 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 19 |
Hb_000788_030 |
0.0890611394 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 20 |
Hb_002272_030 |
0.0902516861 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |