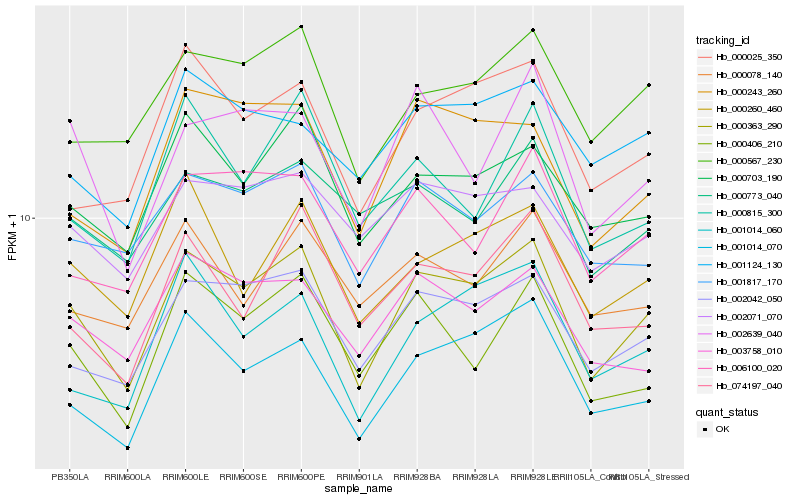
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_290 |
0.0 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 2 |
Hb_002042_050 |
0.0697790296 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001014_070 |
0.0723897707 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 4 |
Hb_000703_190 |
0.0791510615 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000567_230 |
0.0819064015 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 6 |
Hb_001817_170 |
0.0831844758 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 7 |
Hb_000243_260 |
0.0843028453 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000025_350 |
0.0856707696 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003758_010 |
0.0862006323 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000406_210 |
0.0865650965 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 11 |
Hb_000260_460 |
0.0879318567 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 12 |
Hb_002639_040 |
0.0897054506 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 13 |
Hb_002071_070 |
0.0898233377 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001014_060 |
0.0907380435 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 15 |
Hb_006100_020 |
0.0911325658 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 16 |
Hb_074197_040 |
0.0929586611 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 17 |
Hb_001124_130 |
0.0944259084 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 18 |
Hb_000773_040 |
0.0958181465 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000815_300 |
0.0969549408 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 20 |
Hb_000078_140 |
0.0970900477 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |