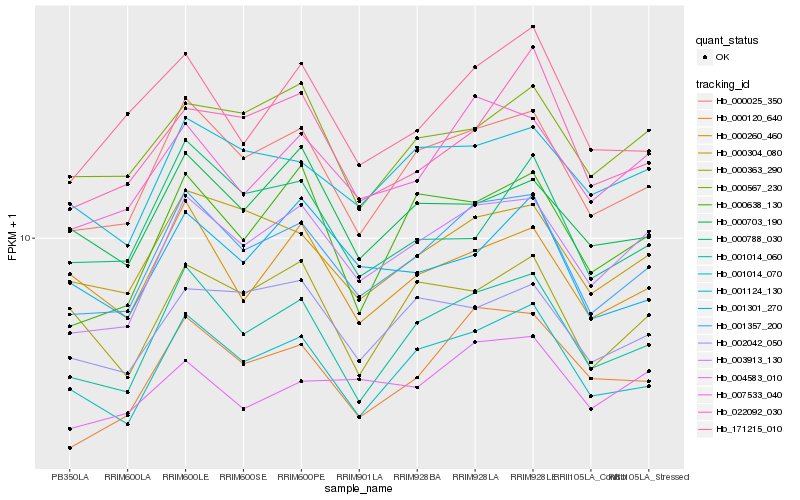
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_200 |
0.0 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_000025_350 |
0.0419500729 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001014_060 |
0.0529343038 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 4 |
Hb_003913_130 |
0.0612449012 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 5 |
Hb_001014_070 |
0.0629894252 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 6 |
Hb_000304_080 |
0.072781046 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007533_040 |
0.0751776524 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 8 |
Hb_000120_640 |
0.0820005381 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001124_130 |
0.0875189115 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 10 |
Hb_171215_010 |
0.0904831525 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 11 |
Hb_002042_050 |
0.091195051 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000260_460 |
0.0925457867 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000638_130 |
0.0943424142 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 14 |
Hb_000567_230 |
0.0986137517 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 15 |
Hb_022092_030 |
0.1007378098 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001301_270 |
0.1020282597 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 17 |
Hb_000788_030 |
0.1025957986 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 18 |
Hb_000703_190 |
0.1026554287 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000363_290 |
0.1028479354 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 20 |
Hb_004583_010 |
0.103542887 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22150, chloroplastic [Jatropha curcas] |