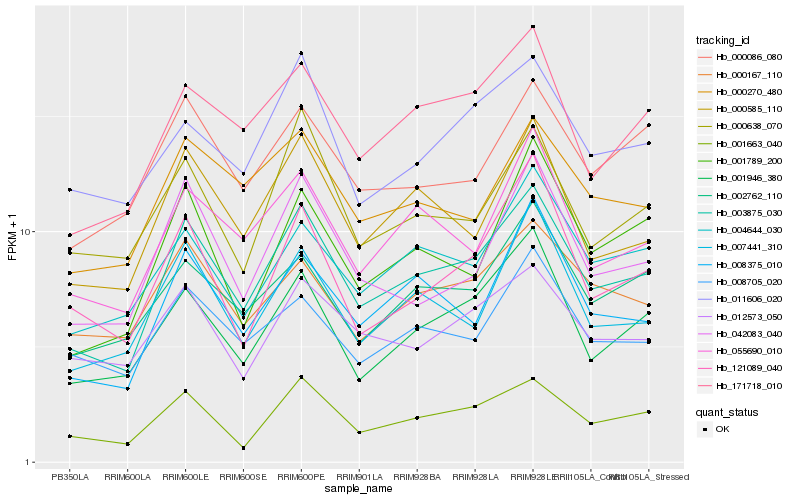
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003875_030 |
0.0 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 2 |
Hb_001663_040 |
0.0840133502 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 3 |
Hb_171718_010 |
0.0855272203 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 4 |
Hb_001946_380 |
0.0869145393 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 5 |
Hb_000585_110 |
0.0895473675 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 6 |
Hb_055690_010 |
0.0937517571 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_008375_010 |
0.0949810082 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 8 |
Hb_042083_040 |
0.0984277621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004644_030 |
0.098577761 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 10 |
Hb_007441_310 |
0.0998160089 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000270_480 |
0.0999006375 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 12 |
Hb_002762_110 |
0.1000545143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011606_020 |
0.1036138427 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 14 |
Hb_000167_110 |
0.1040847028 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_008705_020 |
0.1040874652 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000638_070 |
0.1044709543 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 17 |
Hb_001789_200 |
0.1045237328 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000086_080 |
0.1048137328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012573_050 |
0.1051583204 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 20 |
Hb_121089_040 |
0.1058078766 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |