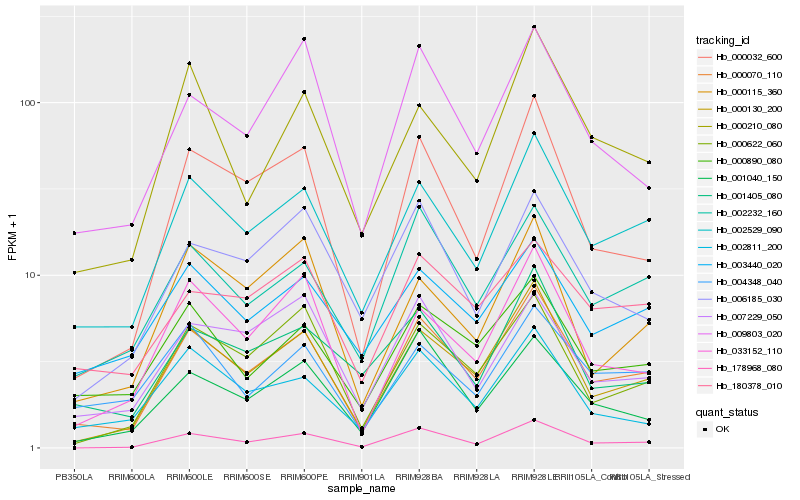
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000070_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000130_200 |
0.0731646389 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 3 |
Hb_002529_090 |
0.0772338003 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_178968_080 |
0.1044155269 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002232_160 |
0.1085462799 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 6 |
Hb_000032_600 |
0.1097852661 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 7 |
Hb_000890_080 |
0.1109543982 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 8 |
Hb_000622_060 |
0.1157075669 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_033152_110 |
0.1221346793 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 10 |
Hb_001040_150 |
0.1263564973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009803_020 |
0.1271768759 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 12 |
Hb_180378_010 |
0.1300241024 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 13 |
Hb_003440_020 |
0.1314773582 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_002811_200 |
0.1316021891 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 15 |
Hb_000210_080 |
0.1337166865 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_004348_040 |
0.1352586007 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 17 |
Hb_001405_080 |
0.1354791627 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 18 |
Hb_007229_050 |
0.1368037114 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000115_360 |
0.1392880066 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 20 |
Hb_006185_030 |
0.1404095694 |
- |
- |
threonine synthase, putative [Ricinus communis] |