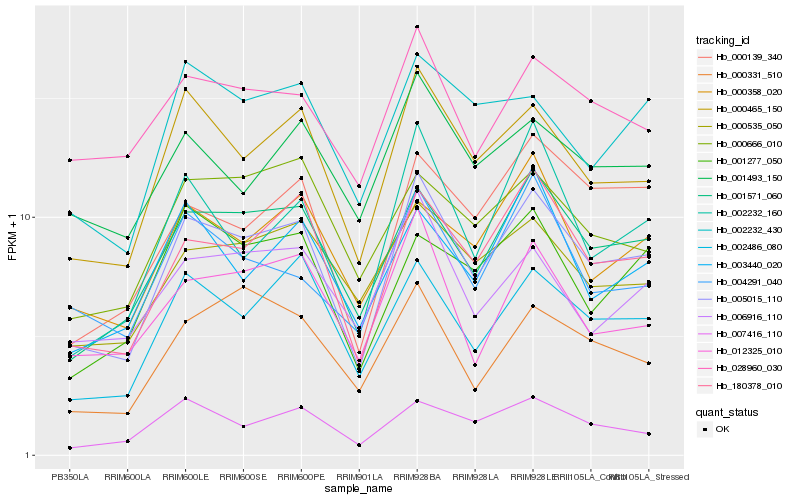
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005015_110 |
0.0 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 2 |
Hb_000465_150 |
0.0799699184 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 3 |
Hb_180378_010 |
0.0810653507 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 4 |
Hb_001571_060 |
0.0831993631 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 5 |
Hb_002486_080 |
0.0947598371 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 6 |
Hb_000139_340 |
0.1093872676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007416_110 |
0.1095588803 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002232_160 |
0.1111242394 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 9 |
Hb_000535_050 |
0.1150985621 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 10 |
Hb_012325_010 |
0.1167952599 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 11 |
Hb_028960_030 |
0.1178539936 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 12 |
Hb_006916_110 |
0.1202443459 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000358_020 |
0.121144135 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 14 |
Hb_001493_150 |
0.1219870127 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 15 |
Hb_004291_040 |
0.1227362144 |
- |
- |
hexokinase [Manihot esculenta] |
| 16 |
Hb_002232_430 |
0.1229565201 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001277_050 |
0.1229818346 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000331_510 |
0.1239280047 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 19 |
Hb_000666_010 |
0.1242798373 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003440_020 |
0.1244523883 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |