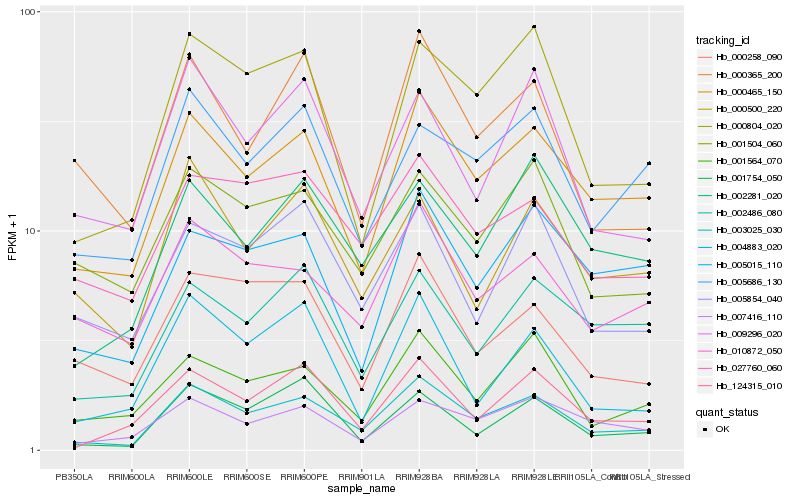
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003025_030 |
0.0 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 2 |
Hb_000465_150 |
0.0976554062 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 3 |
Hb_000804_020 |
0.1169571355 |
- |
- |
PREDICTED: aconitate hydratase, cytoplasmic [Jatropha curcas] |
| 4 |
Hb_124315_010 |
0.1293085205 |
- |
- |
PREDICTED: uncharacterized protein LOC105650413 [Jatropha curcas] |
| 5 |
Hb_002281_020 |
0.1303017804 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 6 |
Hb_001564_070 |
0.1306188822 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 7 |
Hb_007416_110 |
0.1344011434 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010872_050 |
0.1362155226 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 9 |
Hb_027760_060 |
0.137240722 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 10 |
Hb_005686_130 |
0.1375529503 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 11 |
Hb_000365_200 |
0.137793286 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 12 |
Hb_001754_050 |
0.1397201058 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_009296_020 |
0.140240293 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_002486_080 |
0.1411460561 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 15 |
Hb_004883_020 |
0.1416094425 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 16 |
Hb_000500_220 |
0.1416587765 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 17 |
Hb_001504_060 |
0.1427845838 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 18 |
Hb_005854_040 |
0.1435531743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005015_110 |
0.1444161169 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 20 |
Hb_000258_090 |
0.1445496823 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |