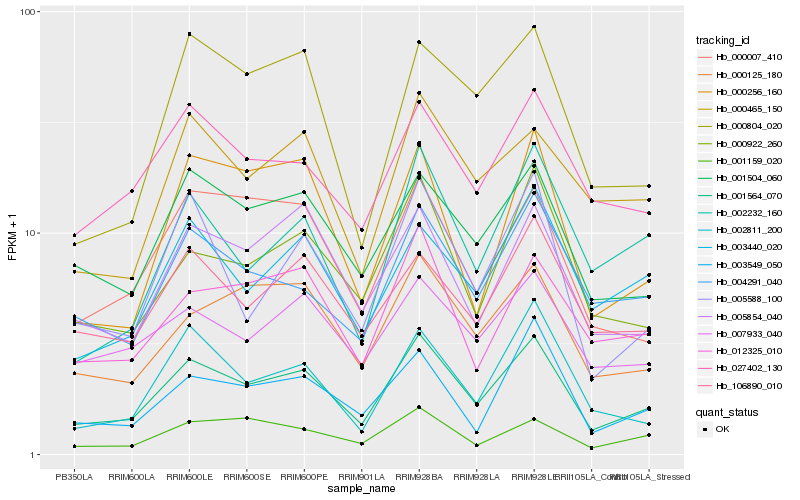
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001564_070 |
0.0 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 2 |
Hb_001504_060 |
0.1016926639 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 3 |
Hb_000125_180 |
0.1064493026 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 4 |
Hb_000922_260 |
0.1077469513 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 5 |
Hb_005854_040 |
0.1103850579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004291_040 |
0.1124038873 |
- |
- |
hexokinase [Manihot esculenta] |
| 7 |
Hb_012325_010 |
0.1125396363 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 8 |
Hb_027402_130 |
0.112842331 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_000007_410 |
0.1133529548 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 10 |
Hb_003440_020 |
0.1139775492 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_005588_100 |
0.1140220128 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 12 |
Hb_106890_010 |
0.1148720469 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_007933_040 |
0.1162204744 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003549_050 |
0.1177862086 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 15 |
Hb_000256_160 |
0.1190833072 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 16 |
Hb_001159_020 |
0.1195507089 |
- |
- |
- |
| 17 |
Hb_000465_150 |
0.1204073082 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 18 |
Hb_002811_200 |
0.1209642313 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 19 |
Hb_000804_020 |
0.1218379509 |
- |
- |
PREDICTED: aconitate hydratase, cytoplasmic [Jatropha curcas] |
| 20 |
Hb_002232_160 |
0.1220107306 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |