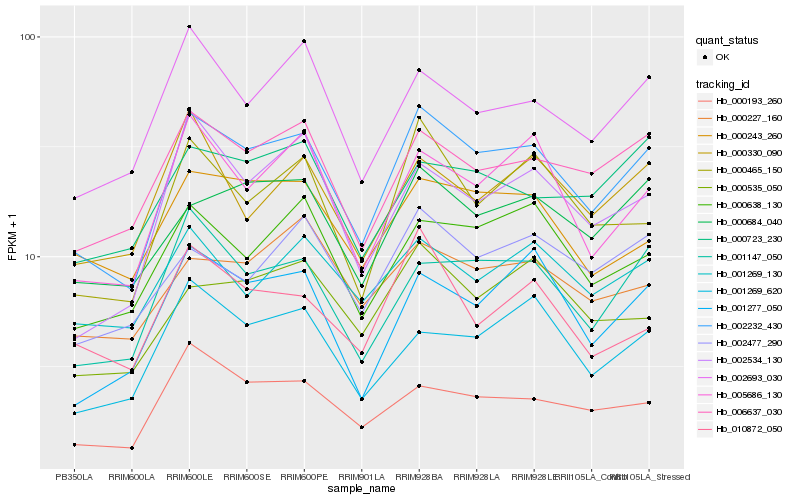
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_430 |
0.0 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 2 |
Hb_006637_030 |
0.0925289625 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 3 |
Hb_001147_050 |
0.0972353602 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000684_040 |
0.0986736234 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 5 |
Hb_000465_150 |
0.1001138516 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 6 |
Hb_005686_130 |
0.1014044982 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 7 |
Hb_000638_130 |
0.1050698927 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 8 |
Hb_002693_030 |
0.1061343324 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 9 |
Hb_001277_050 |
0.1062516652 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000193_260 |
0.1076871262 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 11 |
Hb_000243_260 |
0.1077514238 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000535_050 |
0.1097773102 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 13 |
Hb_002477_290 |
0.109971672 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_001269_620 |
0.111795518 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 15 |
Hb_001269_130 |
0.1129813396 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 16 |
Hb_002534_130 |
0.1144780973 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 17 |
Hb_000723_230 |
0.1172705937 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 18 |
Hb_000330_090 |
0.1177764357 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000227_160 |
0.1186668064 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 20 |
Hb_010872_050 |
0.1188256199 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |