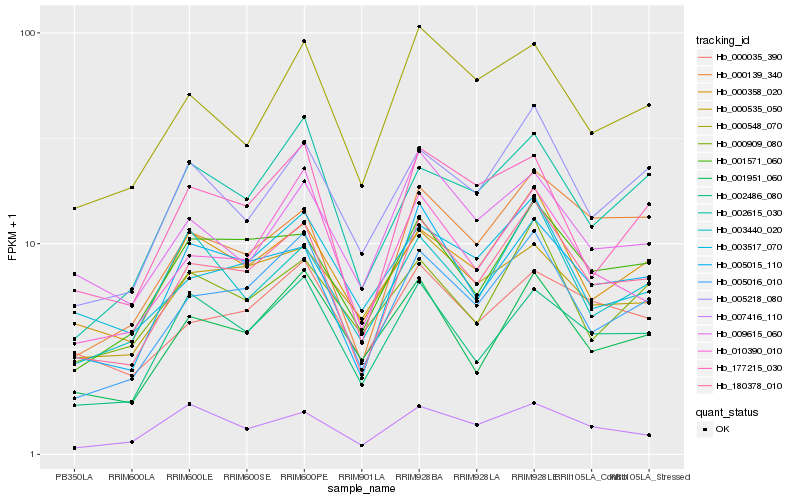
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_180378_010 |
0.0 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 2 |
Hb_005016_010 |
0.0682542443 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 3 |
Hb_005015_110 |
0.0810653507 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 4 |
Hb_001571_060 |
0.0903039561 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 5 |
Hb_000139_340 |
0.0907678614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000358_020 |
0.0927619484 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 7 |
Hb_005218_080 |
0.096978525 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002486_080 |
0.100060766 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 9 |
Hb_010390_010 |
0.1033566743 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 10 |
Hb_000548_070 |
0.1041601092 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 11 |
Hb_001951_060 |
0.1049329367 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 12 |
Hb_177215_030 |
0.104934259 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_000035_390 |
0.1078481561 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 14 |
Hb_002615_030 |
0.1108908531 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 15 |
Hb_003440_020 |
0.1131461205 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_003517_070 |
0.1144515154 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 17 |
Hb_009615_060 |
0.115074638 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 18 |
Hb_007416_110 |
0.1155085374 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000535_050 |
0.1160308955 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 20 |
Hb_000909_080 |
0.1177252339 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |