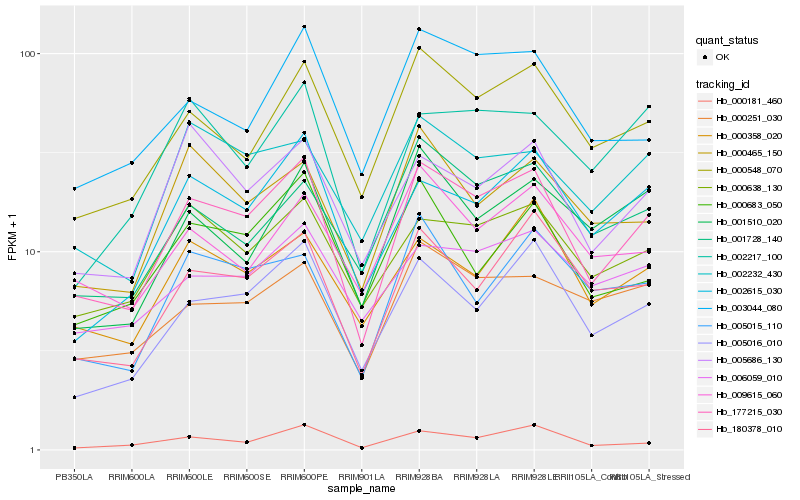
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_177215_030 |
0.0 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_000548_070 |
0.0987356441 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_005016_010 |
0.0996090999 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 4 |
Hb_180378_010 |
0.104934259 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 5 |
Hb_002615_030 |
0.1101150417 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 6 |
Hb_000638_130 |
0.1116691855 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 7 |
Hb_000181_460 |
0.1158151979 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 8 |
Hb_005686_130 |
0.1180417293 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 9 |
Hb_003044_080 |
0.1181070915 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_000251_030 |
0.1205938903 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001728_140 |
0.1219730559 |
- |
- |
- |
| 12 |
Hb_000358_020 |
0.1220007863 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 13 |
Hb_000465_150 |
0.1225531338 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 14 |
Hb_002232_430 |
0.1229188287 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000683_050 |
0.1251663319 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 16 |
Hb_006059_010 |
0.125381413 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 17 |
Hb_009615_060 |
0.1266130736 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 18 |
Hb_005015_110 |
0.1271708191 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 19 |
Hb_002217_100 |
0.127365429 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_001510_020 |
0.1275127938 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |