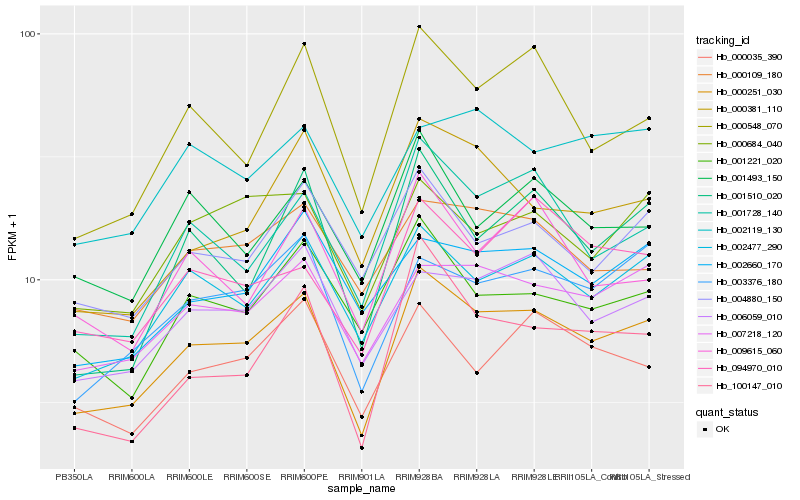
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000251_030 |
0.0 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_002477_290 |
0.0787449832 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_007218_120 |
0.0888990802 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001728_140 |
0.091375811 |
- |
- |
- |
| 5 |
Hb_003376_180 |
0.0919661567 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 6 |
Hb_006059_010 |
0.0958019033 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 7 |
Hb_000548_070 |
0.0987335092 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_004880_150 |
0.1000472208 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 9 |
Hb_000684_040 |
0.1002979797 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 10 |
Hb_000035_390 |
0.1043908148 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 11 |
Hb_000109_180 |
0.1044038612 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 12 |
Hb_001510_020 |
0.1046257984 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 13 |
Hb_001221_020 |
0.1049438286 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 14 |
Hb_001493_150 |
0.1052303274 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 15 |
Hb_002660_170 |
0.1056020957 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 16 |
Hb_100147_010 |
0.1056457075 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 17 |
Hb_000381_110 |
0.1075180201 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_094970_010 |
0.1094441099 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002119_130 |
0.1104923413 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 20 |
Hb_009615_060 |
0.11114028 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |