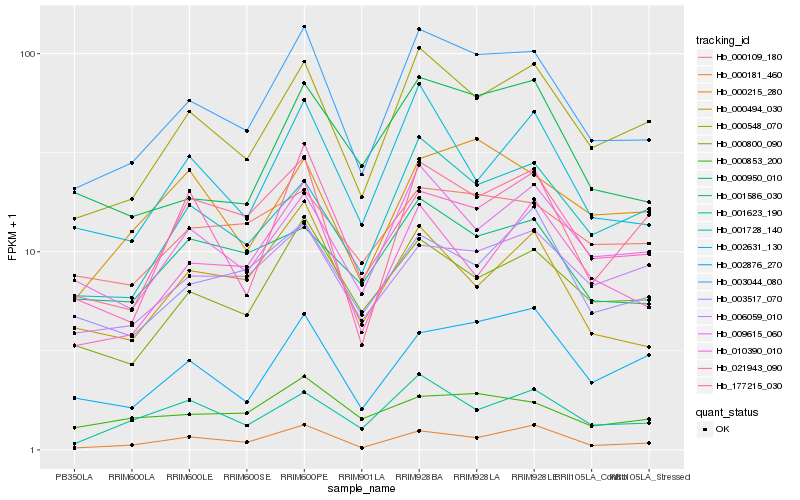
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003044_080 |
0.0 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 2 |
Hb_000548_070 |
0.0779328123 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_000800_090 |
0.101577027 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 4 |
Hb_009615_060 |
0.104850611 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 5 |
Hb_001728_140 |
0.1090669675 |
- |
- |
- |
| 6 |
Hb_002631_130 |
0.1120141764 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 7 |
Hb_000950_010 |
0.1124137366 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 8 |
Hb_177215_030 |
0.1181070915 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_002876_270 |
0.1190018127 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000494_030 |
0.1208203425 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 11 |
Hb_010390_010 |
0.1211274958 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 12 |
Hb_001623_190 |
0.1229887867 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 13 |
Hb_000853_200 |
0.1230322313 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 14 |
Hb_000181_460 |
0.1230381554 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 15 |
Hb_003517_070 |
0.1243089189 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 16 |
Hb_021943_090 |
0.12472816 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 17 |
Hb_000109_180 |
0.1250490129 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 18 |
Hb_006059_010 |
0.1251443128 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 19 |
Hb_001586_030 |
0.1251642964 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 20 |
Hb_000215_280 |
0.127463075 |
- |
- |
Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor, putative [Ricinus communis] |