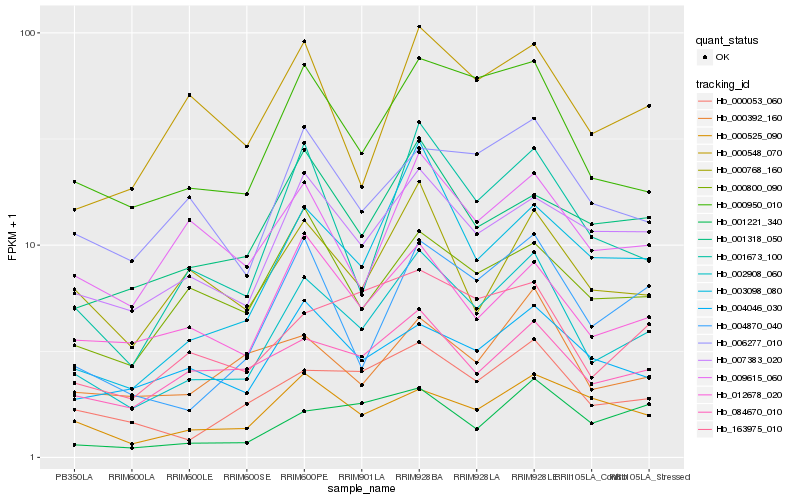
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002908_060 |
0.0 |
- |
- |
unnamed protein product, partial [Coffea canephora] |
| 2 |
Hb_000950_010 |
0.1097887495 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 3 |
Hb_000053_060 |
0.1179216673 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |
| 4 |
Hb_001673_100 |
0.121852944 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 5 |
Hb_004870_040 |
0.1261696412 |
- |
- |
- |
| 6 |
Hb_007383_020 |
0.130066919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003098_080 |
0.1358535627 |
- |
- |
- |
| 8 |
Hb_163975_010 |
0.1365177808 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_012678_020 |
0.1407926879 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 10 |
Hb_084670_010 |
0.1444667431 |
- |
- |
PREDICTED: thiamine biosynthetic bifunctional enzyme TH1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_006277_010 |
0.1449142586 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 12 |
Hb_000525_090 |
0.1491916081 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 13 |
Hb_000392_160 |
0.1499634159 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001221_340 |
0.1499776982 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 15 |
Hb_009615_060 |
0.1500256734 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 16 |
Hb_001318_050 |
0.1504461453 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 17 |
Hb_004046_030 |
0.1517060411 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 18 |
Hb_000800_090 |
0.1533370113 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000768_160 |
0.1536172301 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 20 |
Hb_000548_070 |
0.1550699536 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |