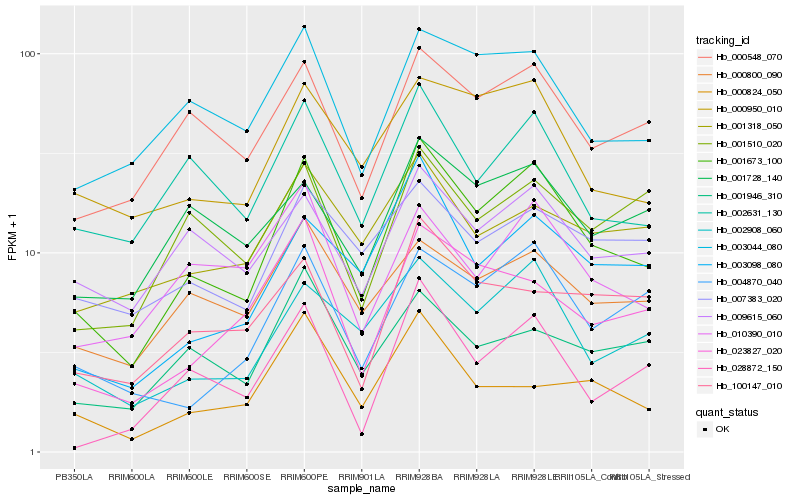
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001673_100 |
0.0 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 2 |
Hb_002908_060 |
0.121852944 |
- |
- |
unnamed protein product, partial [Coffea canephora] |
| 3 |
Hb_002631_130 |
0.1329959102 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 4 |
Hb_000548_070 |
0.1352305165 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 5 |
Hb_003098_080 |
0.1367481227 |
- |
- |
- |
| 6 |
Hb_000950_010 |
0.1380015692 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 7 |
Hb_004870_040 |
0.1381782316 |
- |
- |
- |
| 8 |
Hb_003044_080 |
0.1411661424 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 9 |
Hb_009615_060 |
0.1433364281 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 10 |
Hb_023827_020 |
0.1440675645 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 11 |
Hb_000800_090 |
0.1495031965 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 12 |
Hb_010390_010 |
0.1499627698 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 13 |
Hb_100147_010 |
0.1539704881 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 14 |
Hb_001728_140 |
0.1540223039 |
- |
- |
- |
| 15 |
Hb_007383_020 |
0.1548112425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_028872_150 |
0.1556705019 |
- |
- |
PREDICTED: uncharacterized protein LOC105137493 [Populus euphratica] |
| 17 |
Hb_001946_310 |
0.1573631468 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 18 |
Hb_001318_050 |
0.1577654221 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 19 |
Hb_001510_020 |
0.1595900802 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 20 |
Hb_000824_050 |
0.1642230151 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |