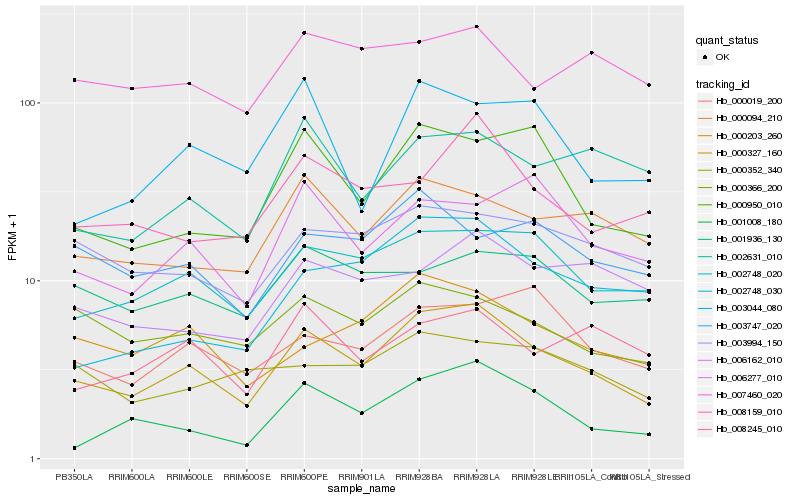
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_160 |
0.0 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 2 |
Hb_002748_020 |
0.1225939485 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 3 |
Hb_000950_010 |
0.132545925 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 4 |
Hb_000203_260 |
0.1349365125 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like [Jatropha curcas] |
| 5 |
Hb_000094_210 |
0.136930836 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 6 |
Hb_000366_200 |
0.1428957403 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 7 |
Hb_003994_150 |
0.1440282012 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 [Jatropha curcas] |
| 8 |
Hb_001008_180 |
0.1444540814 |
- |
- |
- |
| 9 |
Hb_008159_010 |
0.1452559724 |
- |
- |
PREDICTED: 66 kDa stress protein [Jatropha curcas] |
| 10 |
Hb_000019_200 |
0.146933474 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 11 |
Hb_006277_010 |
0.1511938813 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 12 |
Hb_002748_030 |
0.1542743297 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 13 |
Hb_008245_010 |
0.1550191636 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |
| 14 |
Hb_003044_080 |
0.155693441 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 15 |
Hb_003747_020 |
0.1564981179 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X1 [Jatropha curcas] |
| 16 |
Hb_002631_010 |
0.1567159046 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 17 |
Hb_000352_340 |
0.158028837 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 18 |
Hb_001936_130 |
0.1595965533 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 19 |
Hb_006162_010 |
0.1602720003 |
- |
- |
CAZy families GH95 protein, partial [uncultured Chitinophaga sp.] |
| 20 |
Hb_007460_020 |
0.1614240314 |
- |
- |
PREDICTED: guanosine nucleotide diphosphate dissociation inhibitor 1 [Jatropha curcas] |