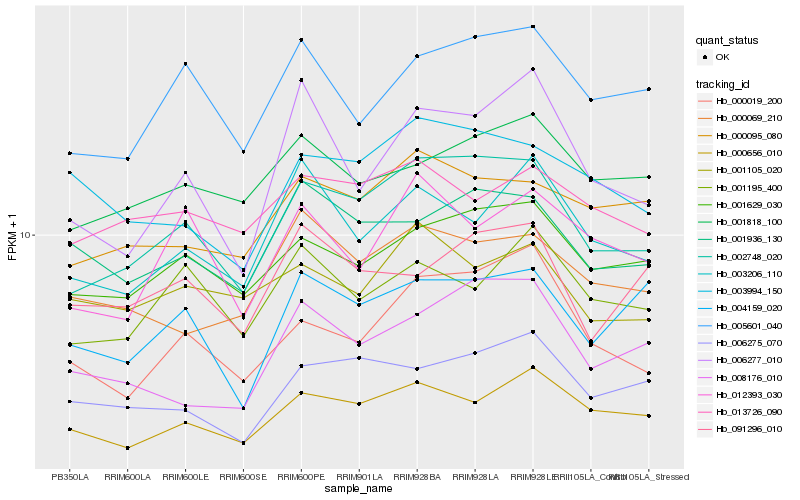
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002748_020 |
0.0 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 2 |
Hb_001629_030 |
0.0833572894 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 3 |
Hb_000019_200 |
0.0921853293 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_005601_040 |
0.0922984559 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000095_080 |
0.0945371738 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 6 |
Hb_000656_010 |
0.0960923381 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006275_070 |
0.1002980444 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 8 |
Hb_001936_130 |
0.1009318709 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 9 |
Hb_001818_100 |
0.1040051184 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 10 |
Hb_004159_020 |
0.104023315 |
- |
- |
Putative dihydrodipicolinate reductase 2, chloroplastic [Aegilops tauschii] |
| 11 |
Hb_008176_010 |
0.1052707678 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20090 [Jatropha curcas] |
| 12 |
Hb_003206_110 |
0.1063635499 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 13 |
Hb_000069_210 |
0.1069569589 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 14 |
Hb_012393_030 |
0.1074376233 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 15 |
Hb_003994_150 |
0.1081742441 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 [Jatropha curcas] |
| 16 |
Hb_006277_010 |
0.1084533084 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 17 |
Hb_001195_400 |
0.1085337466 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 18 |
Hb_013726_090 |
0.1096991151 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 19 |
Hb_001105_020 |
0.1110499501 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 20 |
Hb_091296_010 |
0.1114737345 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |