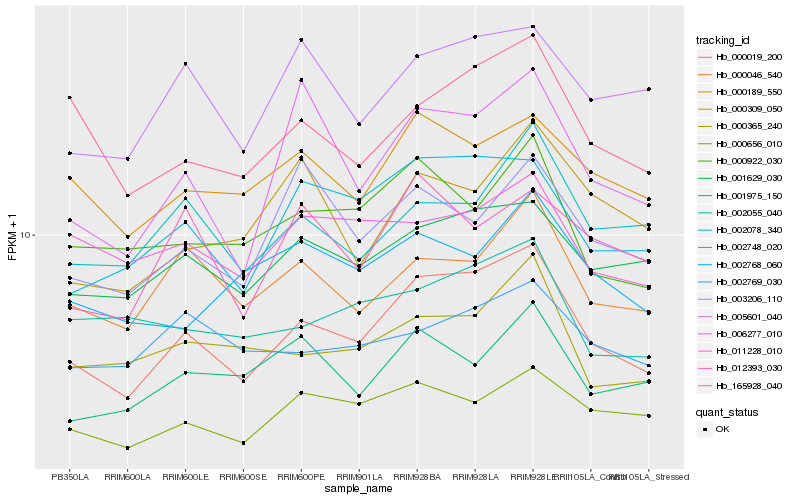
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000019_200 |
0.0 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 2 |
Hb_002748_020 |
0.0921853293 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 3 |
Hb_001629_030 |
0.0991564741 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 4 |
Hb_165928_040 |
0.1013421496 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase component HRD3A [Jatropha curcas] |
| 5 |
Hb_000046_540 |
0.104611599 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 6 |
Hb_000189_550 |
0.1046860851 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 7 |
Hb_006277_010 |
0.1064434726 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 8 |
Hb_000656_010 |
0.1075541923 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005601_040 |
0.1086175527 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 10 |
Hb_002078_340 |
0.1101529108 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000365_240 |
0.1103610111 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 12 |
Hb_000922_030 |
0.1105548398 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002055_040 |
0.1170008987 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 14 |
Hb_001975_150 |
0.1193623841 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 15 |
Hb_012393_030 |
0.119562174 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 16 |
Hb_003206_110 |
0.1198244881 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 17 |
Hb_002768_060 |
0.1201495818 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011228_010 |
0.1220184515 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000309_050 |
0.1221572458 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002769_030 |
0.122553912 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |