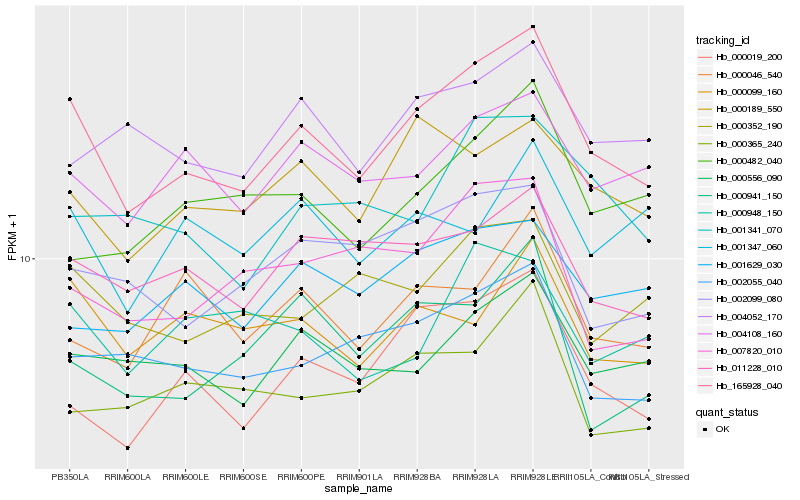
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_165928_040 |
0.0 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase component HRD3A [Jatropha curcas] |
| 2 |
Hb_000556_090 |
0.0995397702 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000019_200 |
0.1013421496 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000189_550 |
0.1015222862 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 5 |
Hb_011228_010 |
0.1016906405 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_002055_040 |
0.1024582173 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 7 |
Hb_004108_160 |
0.1029974488 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 8 |
Hb_000352_190 |
0.1054710276 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 41 [Jatropha curcas] |
| 9 |
Hb_000099_160 |
0.1071620354 |
- |
- |
PREDICTED: potassium transporter 3 isoform X2 [Populus euphratica] |
| 10 |
Hb_002099_080 |
0.1080790894 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 11 |
Hb_001341_070 |
0.108409194 |
- |
- |
PREDICTED: uncharacterized protein LOC105628138 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004052_170 |
0.1106038091 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001629_030 |
0.1128654031 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 14 |
Hb_007820_010 |
0.1147010765 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: protein FLOWERING LOCUS D [Jatropha curcas] |
| 15 |
Hb_000046_540 |
0.1156742768 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 16 |
Hb_000365_240 |
0.1168811741 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 17 |
Hb_000948_150 |
0.1185021409 |
- |
- |
PREDICTED: heme-binding-like protein At3g10130, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000941_150 |
0.1186143813 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000482_040 |
0.1192183277 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 20 |
Hb_001347_060 |
0.1193147013 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |