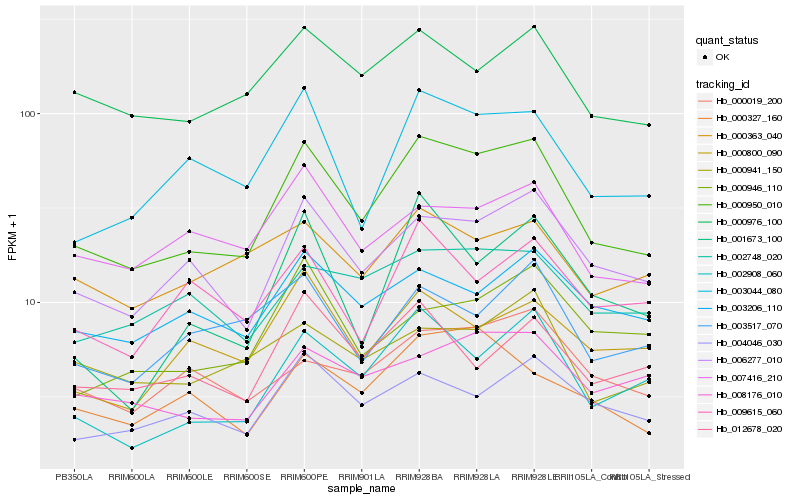
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000950_010 |
0.0 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 2 |
Hb_006277_010 |
0.101055861 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 3 |
Hb_002908_060 |
0.1097887495 |
- |
- |
unnamed protein product, partial [Coffea canephora] |
| 4 |
Hb_003044_080 |
0.1124137366 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 5 |
Hb_000976_100 |
0.1195611236 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 6 |
Hb_004046_030 |
0.1240614721 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 7 |
Hb_000327_160 |
0.132545925 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_007416_210 |
0.1328954758 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 9 |
Hb_003206_110 |
0.1332551193 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 10 |
Hb_008176_010 |
0.133808259 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20090 [Jatropha curcas] |
| 11 |
Hb_002748_020 |
0.1343939959 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 12 |
Hb_000800_090 |
0.1345996121 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000941_150 |
0.1352924704 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000019_200 |
0.1354813785 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 15 |
Hb_012678_020 |
0.1364506687 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000363_040 |
0.1368218709 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 17 |
Hb_003517_070 |
0.1378851742 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 18 |
Hb_001673_100 |
0.1380015692 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 19 |
Hb_000946_110 |
0.1389814723 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 20 |
Hb_009615_060 |
0.1397527441 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |