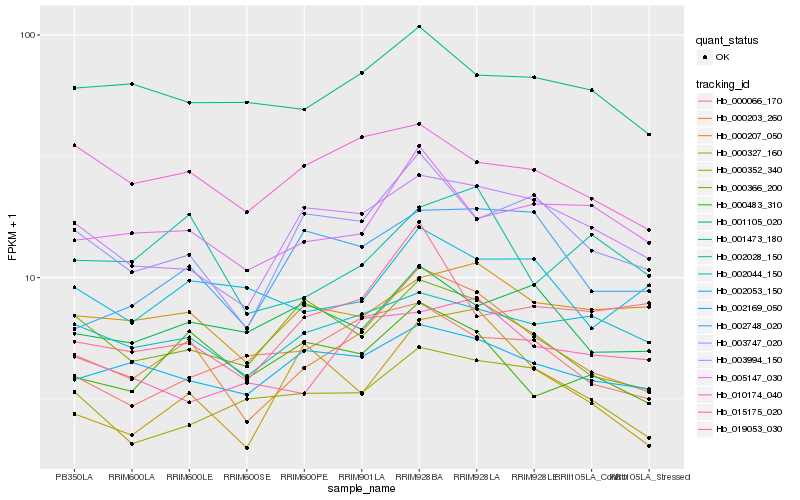
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000203_260 |
0.0 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like [Jatropha curcas] |
| 2 |
Hb_003747_020 |
0.1087419775 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X1 [Jatropha curcas] |
| 3 |
Hb_003994_150 |
0.1194815737 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 [Jatropha curcas] |
| 4 |
Hb_005147_030 |
0.1306567862 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 5 |
Hb_002169_050 |
0.1321908502 |
- |
- |
PREDICTED: inositol 1,3,4-trisphosphate 5/6-kinase 4 [Jatropha curcas] |
| 6 |
Hb_000366_200 |
0.1335167695 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 7 |
Hb_002748_020 |
0.134071997 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 8 |
Hb_000483_310 |
0.1344227269 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4-like [Jatropha curcas] |
| 9 |
Hb_002044_150 |
0.1345506256 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 10 |
Hb_000327_160 |
0.1349365125 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000207_050 |
0.1358667874 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002028_150 |
0.1362487978 |
- |
- |
PREDICTED: hepatocyte growth factor-regulated tyrosine kinase substrate [Jatropha curcas] |
| 13 |
Hb_010174_040 |
0.1380388591 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 14 |
Hb_001473_180 |
0.1381760883 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 15 |
Hb_000352_340 |
0.1400460787 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 16 |
Hb_015175_020 |
0.1409014478 |
- |
- |
hypothetical protein JCGZ_14452 [Jatropha curcas] |
| 17 |
Hb_019053_030 |
0.1412582241 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 18 |
Hb_002053_150 |
0.1430362978 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 19 |
Hb_001105_020 |
0.1431311258 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 20 |
Hb_000066_170 |
0.1454975135 |
- |
- |
PREDICTED: phosphatidylinositol-3-phosphatase myotubularin-1 isoform X2 [Jatropha curcas] |