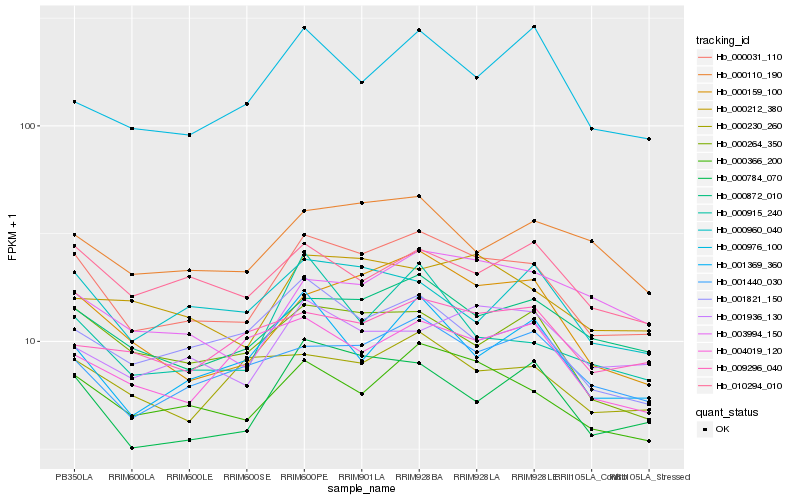
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_110 |
0.0 |
- |
- |
Coatomer, beta subunit isoform 1 [Theobroma cacao] |
| 2 |
Hb_000366_200 |
0.0724734016 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 3 |
Hb_000159_100 |
0.0794778069 |
- |
- |
PREDICTED: uncharacterized protein At4g19900 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000872_010 |
0.0799546794 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 5 |
Hb_000784_070 |
0.0816267749 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000264_350 |
0.093527054 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 7 |
Hb_001440_030 |
0.0939712081 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 8 |
Hb_001821_150 |
0.0973162958 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 9 |
Hb_000212_380 |
0.0985369349 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000110_190 |
0.1004287497 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 11 |
Hb_001369_360 |
0.1019069669 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 12 |
Hb_010294_010 |
0.1059842041 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 13 |
Hb_001936_130 |
0.1064369815 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 14 |
Hb_000915_240 |
0.1069465195 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_003994_150 |
0.1069994699 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 [Jatropha curcas] |
| 16 |
Hb_000960_040 |
0.1071758798 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 17 |
Hb_000230_260 |
0.1071961342 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 18 |
Hb_004019_120 |
0.1078323796 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 19 |
Hb_009296_040 |
0.1080715949 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 20 |
Hb_000976_100 |
0.1086462708 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |