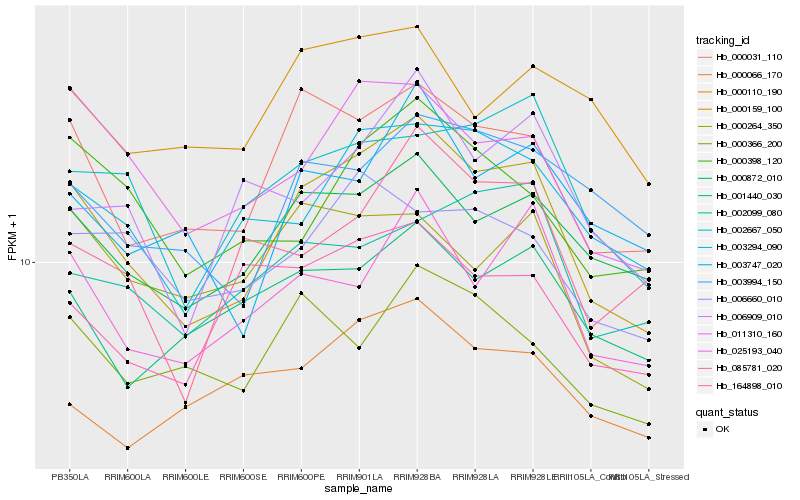
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000159_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g19900 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000031_110 |
0.0794778069 |
- |
- |
Coatomer, beta subunit isoform 1 [Theobroma cacao] |
| 3 |
Hb_000872_010 |
0.0920718796 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 4 |
Hb_011310_160 |
0.0951590713 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 5 |
Hb_003294_090 |
0.0963808546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_025193_040 |
0.0969042143 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 7 |
Hb_002667_050 |
0.1059484592 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 8 |
Hb_000398_120 |
0.1090003702 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3-like isoform X4 [Gossypium raimondii] |
| 9 |
Hb_003747_020 |
0.1105875049 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X1 [Jatropha curcas] |
| 10 |
Hb_003994_150 |
0.1118628558 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 [Jatropha curcas] |
| 11 |
Hb_000366_200 |
0.1127716946 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 12 |
Hb_001440_030 |
0.1131393991 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 13 |
Hb_006909_010 |
0.1137321944 |
- |
- |
5-oxoprolinase, putative [Ricinus communis] |
| 14 |
Hb_000264_350 |
0.1150247227 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 15 |
Hb_164898_010 |
0.1161142181 |
- |
- |
Cysteine-rich protein, putative [Theobroma cacao] |
| 16 |
Hb_006660_010 |
0.1167810574 |
- |
- |
PREDICTED: exocyst complex component EXO84C isoform X1 [Jatropha curcas] |
| 17 |
Hb_000110_190 |
0.1170045384 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 18 |
Hb_085781_020 |
0.1176429152 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 19 |
Hb_002099_080 |
0.1179809405 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 20 |
Hb_000066_170 |
0.1212854392 |
- |
- |
PREDICTED: phosphatidylinositol-3-phosphatase myotubularin-1 isoform X2 [Jatropha curcas] |