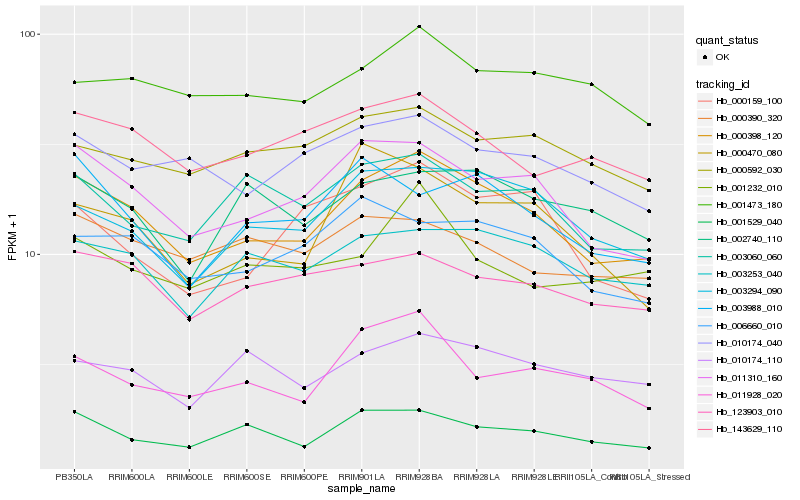
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000398_120 |
0.0 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3-like isoform X4 [Gossypium raimondii] |
| 2 |
Hb_011310_160 |
0.0668131417 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 3 |
Hb_003294_090 |
0.0877348849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001529_040 |
0.0918400943 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 5 |
Hb_003253_040 |
0.0934715258 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 6 |
Hb_006660_010 |
0.0961289191 |
- |
- |
PREDICTED: exocyst complex component EXO84C isoform X1 [Jatropha curcas] |
| 7 |
Hb_143629_110 |
0.0974691575 |
- |
- |
glutamyl-tRNA synthetase, cytoplasmic, putative [Ricinus communis] |
| 8 |
Hb_123903_010 |
0.0997747915 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010174_040 |
0.1010853088 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 10 |
Hb_003060_060 |
0.1026685021 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 11 |
Hb_011928_020 |
0.1045468305 |
- |
- |
something about silencing protein sas10, putative [Ricinus communis] |
| 12 |
Hb_003988_010 |
0.1050004449 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 13 |
Hb_010174_110 |
0.1052396696 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 14 |
Hb_000592_030 |
0.1073143305 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 15 |
Hb_001473_180 |
0.107872442 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 16 |
Hb_002740_110 |
0.10843743 |
- |
- |
PREDICTED: phytochrome A [Jatropha curcas] |
| 17 |
Hb_000159_100 |
0.1090003702 |
- |
- |
PREDICTED: uncharacterized protein At4g19900 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000470_080 |
0.1103538754 |
- |
- |
Caldesmon, putative [Ricinus communis] |
| 19 |
Hb_001232_010 |
0.1111777983 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 20 |
Hb_000390_320 |
0.1116588531 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |