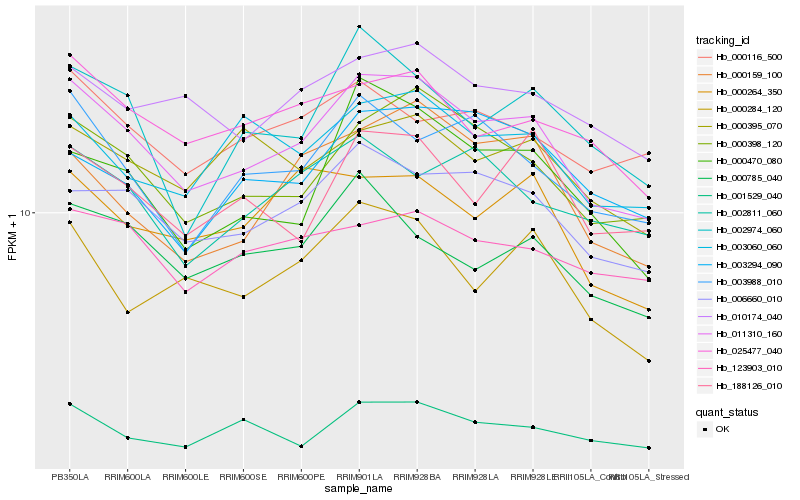
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011310_160 |
0.0 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 2 |
Hb_000398_120 |
0.0668131417 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3-like isoform X4 [Gossypium raimondii] |
| 3 |
Hb_006660_010 |
0.0757292678 |
- |
- |
PREDICTED: exocyst complex component EXO84C isoform X1 [Jatropha curcas] |
| 4 |
Hb_003988_010 |
0.0882818039 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 5 |
Hb_025477_040 |
0.0886922279 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 6 |
Hb_010174_040 |
0.0916690141 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 7 |
Hb_002974_060 |
0.0920808848 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X2 [Jatropha curcas] |
| 8 |
Hb_000785_040 |
0.0936107061 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 9 |
Hb_000159_100 |
0.0951590713 |
- |
- |
PREDICTED: uncharacterized protein At4g19900 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000470_080 |
0.0952542662 |
- |
- |
Caldesmon, putative [Ricinus communis] |
| 11 |
Hb_001529_040 |
0.0952827392 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 12 |
Hb_003060_060 |
0.0956392304 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 13 |
Hb_188126_010 |
0.0958645016 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_123903_010 |
0.0971034209 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000284_120 |
0.0976931927 |
- |
- |
PREDICTED: uncharacterized protein LOC105629656 [Jatropha curcas] |
| 16 |
Hb_000264_350 |
0.0986409127 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 17 |
Hb_002811_060 |
0.1002160062 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 18 |
Hb_000395_070 |
0.1013487482 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000116_500 |
0.1016455252 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 11 [Jatropha curcas] |
| 20 |
Hb_003294_090 |
0.1017493763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |