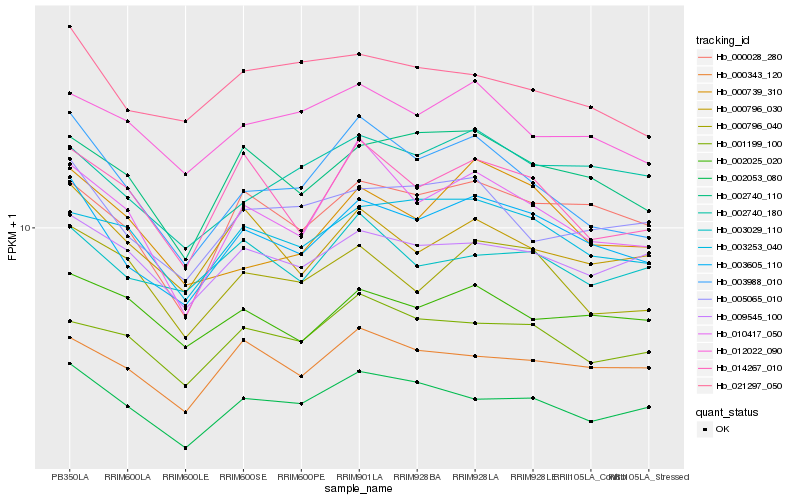
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003605_110 |
0.0 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1D isoform X1 [Jatropha curcas] |
| 2 |
Hb_000028_280 |
0.0668438927 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 3 |
Hb_002740_110 |
0.0695247513 |
- |
- |
PREDICTED: phytochrome A [Jatropha curcas] |
| 4 |
Hb_005065_010 |
0.0777354833 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI11 [Jatropha curcas] |
| 5 |
Hb_003253_040 |
0.079306029 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 6 |
Hb_000343_120 |
0.080179353 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16010 [Jatropha curcas] |
| 7 |
Hb_009545_100 |
0.0805897338 |
- |
- |
PREDICTED: protein OBERON 1 [Jatropha curcas] |
| 8 |
Hb_003988_010 |
0.0809995134 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 9 |
Hb_002740_180 |
0.0813870328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002053_080 |
0.083300052 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21300 [Jatropha curcas] |
| 11 |
Hb_000796_030 |
0.0833946151 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_010417_050 |
0.0836930062 |
- |
- |
PREDICTED: uncharacterized protein LOC105645746 [Jatropha curcas] |
| 13 |
Hb_000796_040 |
0.0845652995 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_014267_010 |
0.0864304394 |
- |
- |
PREDICTED: dnaJ homolog subfamily C GRV2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002025_020 |
0.0872493516 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39710 [Jatropha curcas] |
| 16 |
Hb_012022_090 |
0.0876006681 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Gossypium raimondii] |
| 17 |
Hb_001199_100 |
0.0880912237 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 18 |
Hb_000739_310 |
0.0885824468 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003029_110 |
0.0887137393 |
- |
- |
PREDICTED: uncharacterized protein LOC105640062 isoform X1 [Jatropha curcas] |
| 20 |
Hb_021297_050 |
0.0890080762 |
- |
- |
PREDICTED: signal recognition particle receptor subunit alpha [Jatropha curcas] |