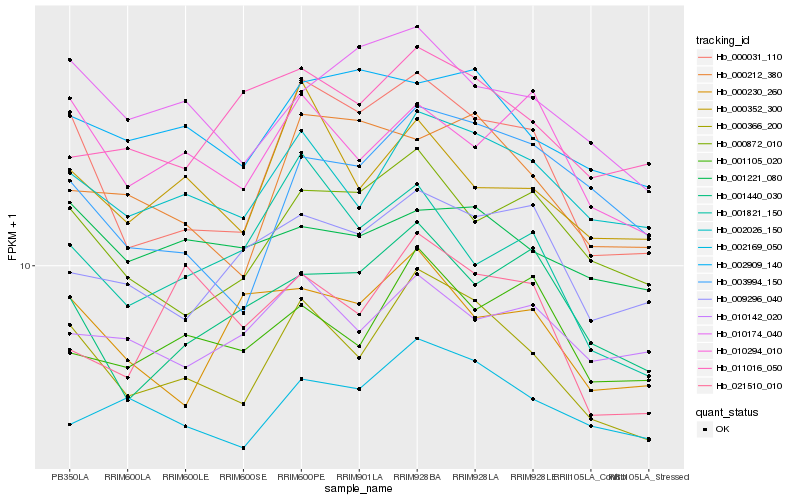
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000366_200 |
0.0 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 2 |
Hb_002026_150 |
0.0659944549 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair helicase XPB1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000031_110 |
0.0724734016 |
- |
- |
Coatomer, beta subunit isoform 1 [Theobroma cacao] |
| 4 |
Hb_000352_300 |
0.0904640297 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 5 |
Hb_002169_050 |
0.0920761859 |
- |
- |
PREDICTED: inositol 1,3,4-trisphosphate 5/6-kinase 4 [Jatropha curcas] |
| 6 |
Hb_010174_040 |
0.0929304801 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 7 |
Hb_002909_140 |
0.0929392794 |
- |
- |
PREDICTED: D-xylose-proton symporter-like 2 [Jatropha curcas] |
| 8 |
Hb_010294_010 |
0.0938546841 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 9 |
Hb_000872_010 |
0.0945245542 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 10 |
Hb_001105_020 |
0.096389877 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 11 |
Hb_000212_380 |
0.0971454036 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 12 |
Hb_003994_150 |
0.1014190958 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 [Jatropha curcas] |
| 13 |
Hb_009296_040 |
0.101487229 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 14 |
Hb_001440_030 |
0.1020169888 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 15 |
Hb_001221_080 |
0.1022542154 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001821_150 |
0.1024276637 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 17 |
Hb_010142_020 |
0.1034323689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011016_050 |
0.1050872299 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000230_260 |
0.105703916 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 20 |
Hb_021510_010 |
0.1058562681 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |