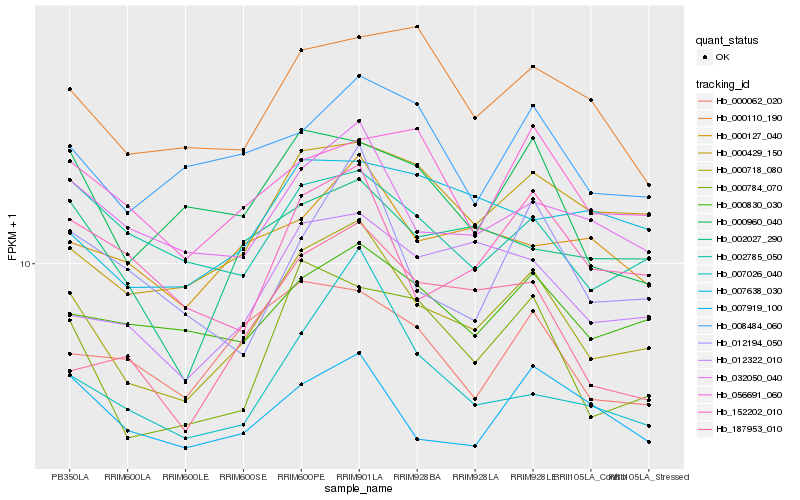
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000718_080 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa005259mg [Prunus persica] |
| 2 |
Hb_000784_070 |
0.0810266835 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_032050_040 |
0.0887646883 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 4 |
Hb_012322_010 |
0.0891956768 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000429_150 |
0.0895587903 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 6 |
Hb_000830_030 |
0.0960366813 |
- |
- |
PREDICTED: uncharacterized protein LOC105644214 [Jatropha curcas] |
| 7 |
Hb_152202_010 |
0.0964802449 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 8 |
Hb_002027_290 |
0.0975074031 |
- |
- |
PREDICTED: NPL4-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000960_040 |
0.0989467267 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 10 |
Hb_000062_020 |
0.0991600367 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_007919_100 |
0.1001022426 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 12 |
Hb_007026_040 |
0.1001363909 |
- |
- |
PREDICTED: chaperonin 60 subunit alpha 2, chloroplastic-like [Eucalyptus grandis] |
| 13 |
Hb_008484_060 |
0.1004231918 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 14 |
Hb_187953_010 |
0.1012976493 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002785_050 |
0.1029140308 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 16 |
Hb_000127_040 |
0.1029909093 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 17 |
Hb_056691_060 |
0.1042766739 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000110_190 |
0.1048330992 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 19 |
Hb_007638_030 |
0.1049658026 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 20 |
Hb_012194_050 |
0.1072679057 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |