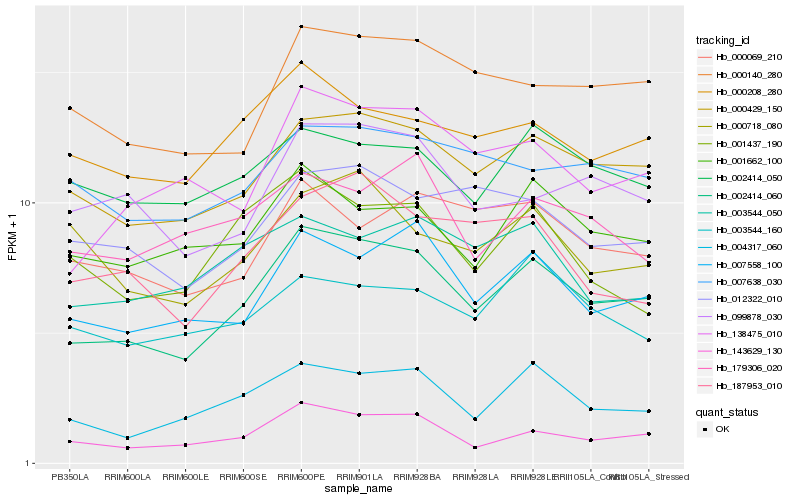
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002414_060 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_E01310 [Eucalyptus grandis] |
| 2 |
Hb_000429_150 |
0.0740487316 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 3 |
Hb_143629_130 |
0.0833363844 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_007558_100 |
0.0852298262 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004317_060 |
0.0879056184 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 6 |
Hb_138475_010 |
0.1001159027 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Populus euphratica] |
| 7 |
Hb_000140_280 |
0.1009791243 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 8 |
Hb_001662_100 |
0.1048123984 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 9 |
Hb_000208_280 |
0.1051614672 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 10 |
Hb_099878_030 |
0.1051688053 |
- |
- |
PREDICTED: coiled-coil domain-containing protein R3HCC1L isoform X4 [Jatropha curcas] |
| 11 |
Hb_012322_010 |
0.1084135129 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_187953_010 |
0.1118386462 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007638_030 |
0.1126286949 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 14 |
Hb_000069_210 |
0.1128574679 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 15 |
Hb_002414_050 |
0.1135306124 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 16 |
Hb_003544_050 |
0.1148702742 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000718_080 |
0.1169365042 |
- |
- |
hypothetical protein PRUPE_ppa005259mg [Prunus persica] |
| 18 |
Hb_003544_160 |
0.1179297969 |
- |
- |
- |
| 19 |
Hb_179306_020 |
0.1179975316 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001437_190 |
0.1180393721 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |