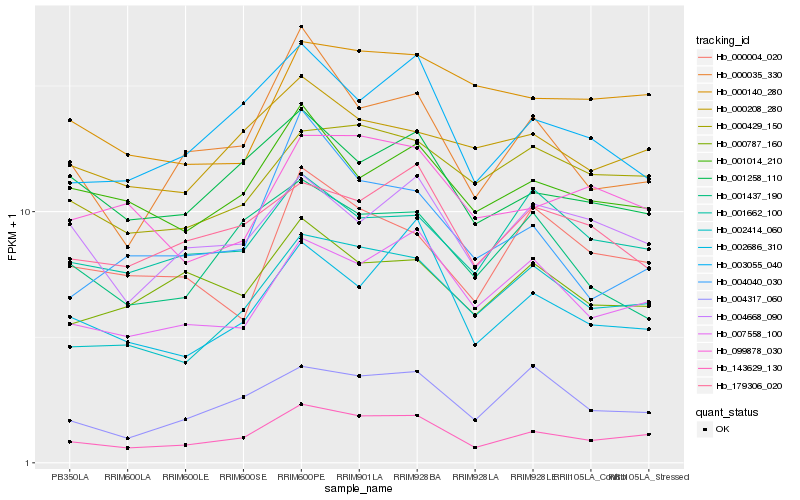
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143629_130 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_002414_060 |
0.0833363844 |
- |
- |
hypothetical protein EUGRSUZ_E01310 [Eucalyptus grandis] |
| 3 |
Hb_007558_100 |
0.0978473676 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000035_330 |
0.0991502197 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |
| 5 |
Hb_003055_040 |
0.1004423044 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002686_310 |
0.1033594069 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 7 |
Hb_004317_060 |
0.1077507988 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 8 |
Hb_001258_110 |
0.1120747224 |
- |
- |
transporter, putative [Ricinus communis] |
| 9 |
Hb_000429_150 |
0.1144707697 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 10 |
Hb_099878_030 |
0.1157665163 |
- |
- |
PREDICTED: coiled-coil domain-containing protein R3HCC1L isoform X4 [Jatropha curcas] |
| 11 |
Hb_001014_210 |
0.1161035508 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 12 |
Hb_179306_020 |
0.1198776319 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000004_020 |
0.1216333036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004040_030 |
0.1221405728 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004668_090 |
0.1231923111 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 16 |
Hb_001662_100 |
0.1231937461 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 17 |
Hb_000208_280 |
0.1251300333 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 18 |
Hb_000787_160 |
0.1255529663 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001437_190 |
0.1266127753 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_000140_280 |
0.1268805612 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |