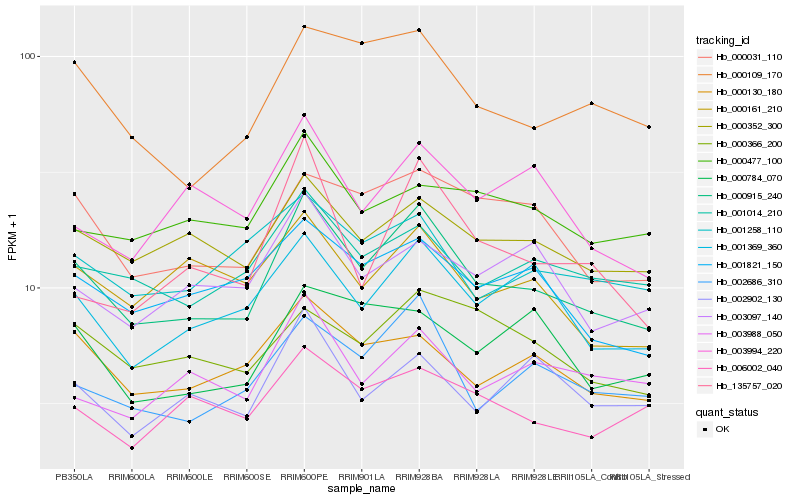
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000915_240 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_002686_310 |
0.0987805189 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 3 |
Hb_001369_360 |
0.0998460199 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 4 |
Hb_000109_170 |
0.1026630975 |
- |
- |
PREDICTED: uncharacterized protein LOC105649868 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000352_300 |
0.1038009217 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 6 |
Hb_000130_180 |
0.1047022019 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_001014_210 |
0.104869841 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 8 |
Hb_000031_110 |
0.1069465195 |
- |
- |
Coatomer, beta subunit isoform 1 [Theobroma cacao] |
| 9 |
Hb_001821_150 |
0.1088786626 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 10 |
Hb_001258_110 |
0.1147148502 |
- |
- |
transporter, putative [Ricinus communis] |
| 11 |
Hb_000161_210 |
0.1182628375 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 12 |
Hb_003097_140 |
0.118639514 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 13 |
Hb_006002_040 |
0.1192411612 |
- |
- |
PREDICTED: uncharacterized protein LOC105638085 [Jatropha curcas] |
| 14 |
Hb_000366_200 |
0.1194995149 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 15 |
Hb_003988_050 |
0.1196476169 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 16 |
Hb_002902_130 |
0.1199986364 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 17 |
Hb_000477_100 |
0.1229710113 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 18 |
Hb_003994_220 |
0.1244969053 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 19 |
Hb_000784_070 |
0.1255552926 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_135757_020 |
0.1256308469 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |