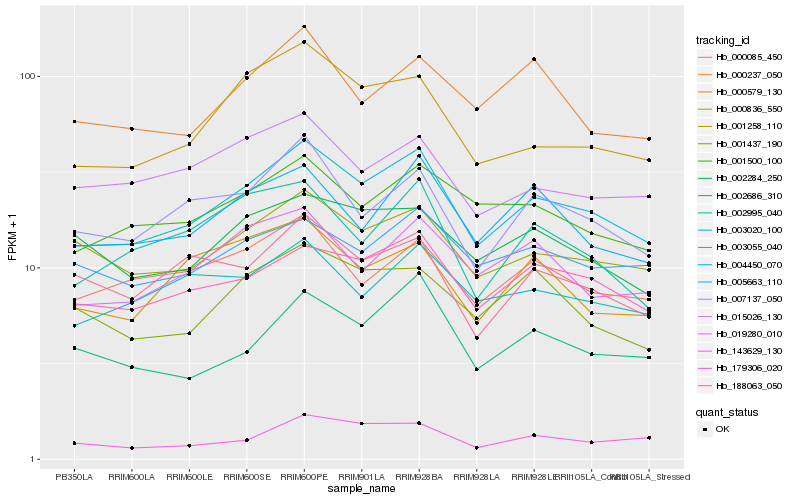
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003055_040 |
0.0 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_179306_020 |
0.0738112723 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001258_110 |
0.0835416768 |
- |
- |
transporter, putative [Ricinus communis] |
| 4 |
Hb_007137_050 |
0.0853066247 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_000836_550 |
0.0868914635 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 6 |
Hb_004450_070 |
0.0883659477 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 7 |
Hb_001500_100 |
0.0894622862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_019280_010 |
0.0905116522 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000085_450 |
0.0937830678 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 10 |
Hb_003020_100 |
0.0958406472 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 11 |
Hb_005663_110 |
0.0968465977 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001437_190 |
0.0974323951 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_002686_310 |
0.0978737195 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 14 |
Hb_000237_050 |
0.1000831322 |
- |
- |
CP2 [Hevea brasiliensis] |
| 15 |
Hb_015026_130 |
0.1004177871 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 16 |
Hb_143629_130 |
0.1004423044 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_002995_040 |
0.1004864174 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002284_250 |
0.1005495425 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 19 |
Hb_188063_050 |
0.102299654 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 20 |
Hb_000579_130 |
0.1023139835 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |